0402
OATP1A1 reporter gene-enhanced magnetic resonance imaging of Triple Negative Breast Cancer in animal models at 3 Tesla1Medical Imaging, Robarts Research Institute, London, ON, Canada, 2Medical Biophysics, University of Western Ontario, London, ON, Canada, 3Ontario Institute for Cancer Research, Toronto, ON, Canada
Synopsis
Preclinical cancer models are invaluable for studying oncogenic pathways and assessing therapies. However, precise detection of small tumours with specificity, sensitivity and high resolution remains challenging. A member of the Organic Anion Transporting Polypeptide 1 (OATP1) family of proteins, called OATP1A1, can take up the clinically-approved, liver-specific paramagnetic agent Gd-EOB-DTPA. Significant increases in spin-lattice relaxation rates were exhibited at 3T by triple negative breast cancer (TNBC) cells engineered to express OATP1A1 and pilot data showed enhancement of OATP1A1-expressing orthotopic TNBC tumours in mice. Our data supports the utility of OATP1A1 for improved MR detection of TNBC tumours in animal models.
Introduction
In studying oncogenesis and assessing therapies in preclinical animal models, detection of small tumors and any metastatic formations in vivo with high conspicuity remains a challenge. Reporter genes are a valuable tool as they encode products that can be detected across time and space, allowing for topographical quantification of engineered cells for longitudinal imaging studies. However, the sensitivity and/or negative contrast of commonly utilized iron-based MR reporter genes is less than ideal. Organic Anion Transporting Polypeptide 1 (OATP1) proteins, expressed endogenously by liver cells, are responsible for T1-shortening in the liver following administration of a clinically-approved liver-specific agent gadolinium ethoxybenzyl diethylene triamine pentaacetic acid (Gd-EOB-DTPA). In 2014, it was shown that ectopic expression of OATP1A1 could be used as a novel MRI reporter gene (1). Moreover, OATP1A1-expressing cells improved bioluminescence imaging (BLI) of luciferase-expressing cells due to increased cell uptake of the luciferase substrate, D-luciferin (2). Our objective is to improve MRI and BLI detectability and intratumoral characterization of luciferase-expressing triple negative breast cancer (TNBC) tumors in mice by co-expressing OATP1A1.Methods
Human (MDA-MB-231) and murine (4T1) TNBC cells were transduced with two separate lentiviruses: the first co-expressing tdTomato and Firefly Luciferase for BLI; and the second co-expressing zsGreen and OATP1A1. Transduced cells were sorted for tdTomato and zsGreen-positive cells. Luciferase lysis assays were performed to measure intracellular luciferase activity and BLI of cells incubated with D-luciferin (150 μg/mL) was performed. Inversion recovery imaging and T1-mapping of cell pellets (5x107 cells) incubated with or without Gd-EOB-DTPA (1.6 mM) for 90 minutes was performed at 3T. MDA-MB-231 cells co-expressing luciferase and OATP1A1 were implanted orthotopically into the mammary fat pad of a pilot female Nu/Nu mouse (n=1). Whole-body BLI, non-contrast MRI, and Gd-EOB-DTPA-enhanced MRI at 3T were acquired. Histological staining and microscopy of tumour sections were performed at endpoint.Results
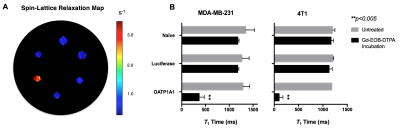
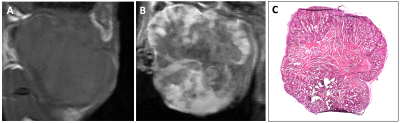
Following lentiviral transduction, fluorescence-activated cell sorting generated populations double-positive for tdTomato and zsGreen with >95% purity (Figure 1A). Immunofluorescence staining confirmed OATP1A1 expression (Figure 1B). MTT proliferation assays showed no difference in relative growth rates between non-transduced and engineered cells (Figure 1C). No difference in intracellular luciferase activity between any of the transduced genotypes was observed (Figure 2A), whereas BLI of intact cells showed significantly higher signals from luciferase/OATP1A1-expressing cells compared to cells expressing luciferase alone (p<0.05; Figures 2B, 2C). This finding was further supported in vivo, as pilot mice implanted orthotopically with bilateral luciferase-expressing and luciferase/OATP1A1-expressing TNBC tumours of equivalent sizes exhibited considerably greater BLI output from their OATP1A1-expressing tumours (Figure 2D). Both human and murine cells displayed significantly increased spin-lattice relaxation rates between OATP1A1-expressing cells incubated with Gd-EOB-DTPA compared to control populations (p<0.05; Figure 3). Interestingly, murine 4T1 cells expressing rat-derived OATP1A1 cells displayed shorter T1 times following incubation than human MDA-MB-231 cells expressing rat-derived OATP1A1, perhaps indicating species-specific post-translational modifications that enhance the transporter’s activity, although this conjecture requires further investigation. Compared to pre-contrast images, heterogeneous intratumoral signal enhancement was observed 4 hours after administration of 0.5 mmol/kg Gd-EOB-DTPA. These areas of enhancement appeared to closely match regions of viable tumour tissue in histological sections (Figure 4), supporting the feasibility of performing highly-resolved and spatially-informative viability MR assays of OATP1A1-engineered cells.Conclusion
Maximizing sensitivity is key to improving our ability to visualize and quantify small tumours in vivo. If the OATP1A1 reporter gene system proves effective, dissemination of this tool paves the path toward improved molecular imaging of engineered cells with high spatial resolution, sensitivity and 3D information. Importantly, MRI reporter gene development has largely focused on iron-based mechanisms that generate continuous negative contrast, which already exists within living organisms. Further, these systems have been strongly suggested to interfere with iron homeostasis and cell phenotype, which may confound biological findings in preclinical research studies (3). Although more work is needed, OATP1A1 could possibly function as an inert positive contrast MRI reporter gene that provides signal only following Gd-EOB-DTPA administration, and concomitantly enhances BLI signal generation and detection depth. Future work will focus on validating this reporter gene in a complete cohort of tumour-bearing animals, and evaluating its use for better detection and characterization of disseminated metastases.Acknowledgements
This work was supported by the Breast Cancer Society of Canada.
References
1. Patrick PS, Hammersleya J, Loizoua L ... Brindle KM (2014). Dual-modality gene reporter for in vivo imaging. PNAS 111(1): 415-20.
2. Patrick PS, Lyons SK, Rodrigues TB, Brindle KM (2014). Oatp1 enhances bioluminescence by acting as a plasma membrane transporter for D-luciferin. Mol Imaging Biol 16(5): 626-34.
3. Pereira SM, Moss D, Williams SR … Taylor A (2015). Overexpression of the MRI reporter genes ferritin and transferrin receptor affect iron homeostasis and produce limited contrast in mesenchymal stem cells. Int J Mol Sci 16(7): 15481–96.
Figures
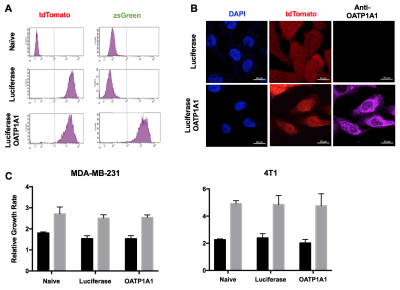
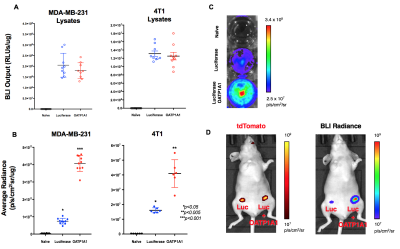
Figure 2. Bioluminescence Imaging. A) BLI output (RLUs/μg±SD) of MDA-MB-231 and 4T1 cell lysates. B) Average radiance (p/s/cm2/μg±SD) of MDA-MB-231 and 4T1 cells in vitro. C) BLI of representative well plate. D) tdTomato FLI and unfiltered BLI of representative mouse implanted orthotopically with bilateral TNBC tumours expressing luciferase (left) or co-expressing luciferase and OATP1A1 (right).