0375
Multiscale multimodal imaging of the mouse cerebral vascular architecture.1Bio-imaging Lab, University of Antwerp, Wilrijk, Belgium, 2Laboratory of Cell Biology and Histology, University of Antwerp, Wilrijk, Belgium, 3Molecular Imaging Center Antwerp, University of Antwerp, Wilrijk, Belgium
Synopsis
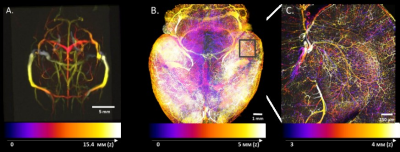
We present a multiscale multimodal atlas of the mouse cerebral vasculature co-registered to the Allen Brain atlas space. To this end, we combined MRI anatomical imaging with the macro vasculature detected with TOF-MRA as well as the micro vasculature acquired from 3D microscopic imaging of a cleared mouse brain. To achieve a better homogeneity, we used a double labeling of the vasculature using isolectin to stain the vascular wall combined with an albumin staining of the blood vessel lumen. The atlas provides a reference space across multiple imaging modalities and a link to other functional, anatomical and genetic information.
Purpose: Vascular abnormalities are hallmark comorbidities of a variety of neuropathologies such as Alzheimer’s disease, stroke, and brain tumors. Hence, in-depth knowledge about the cerebral vascular architecture and its alterations in disease are quintessential. Importantly, a number of brain functional imaging modalities such as functional magnetic resonance imaging (fMRI), functional Ultrasound (fUS), optical imaging of intrinsic signals, and others, also greatly depend on the cerebral vasculature and its correspondence to other functional segmentations of the brain. Here, we used a multi-scale approach combining anatomical MRI and time of flight magnetic resonance angiography (TOF-MRA) with high-resolution whole brain microscopic imaging data on the same animal in order to create a high resolution vascular atlas that could be co-registered to the Allen Brain space providing a link to additional functional, anatomical and genetic information. Further, this atlas can be used as a potential reference for combining information across imaging modalities.
Method: Male C57BL/6 (N=6) mice of 12 weeks old were used throughout the entire study. Animals were anesthetized using 2% isoflurane and were scanned on a 9.4T MRI scanner (Bruker BioSpec, Germany). MRI data acquisition included 3D-T2 anatomical scan (TE/TR 40/2500 ms, isotropic voxel size of 0.078 mm³) followed by three 2D (TOF-MRA) scans orientated in orthogonal directions along the main acquisition axes (TE/TR 3/18, Flip angle 80°, voxel size of 0.063x0.063x0.3 mm) to detect the macro vasculature in each imaging plane. All scans were co-registered to the Allen Brain atlas space using advanced normalization tools (ANTs). The three 2D-TOF scans were combined on a subject-by-subject basis by using a voxel wise maximum intensity projection (MIP). Afterwards, MIPs of each subject were averaged across all subjects to create a template. The micro-vasculature was acquired from iDISCO-cleared mouse brains using a new double labeling technique combining Isolectin staining of the vessel wall with an albumin blood vessel lumen staining. Mice received an intravenous injection with 75 µl (1 mg/ml) as well as a transcardial injection 25 µl (1 mg/ml) of Isolectin GS-IB4 conjugated with Alexa Fluor-647 to label the cerebral vasculature. Subsequently, mice were perfused with 4% PFA at 4°C. This was followed by a perfusion of 10 ml of albumin-FITC conjugate dissolved in 2% gelatin for an additional luminal staining of the blood vessels. Mice were put head-down in ice water for 30 minutes to solidify the gel. Brains were dissected and post-fixed for 24 hours in 4% PFA and iDISCO-cleared. Cleared samples were imaged in a sagittal orientation on a light sheet microscope (Ultramicroscope II, LaVision Biotec) equipped with a sCMOS camera (Andor Neo) and LED lasers (488/561/640 nm). Scans were made at 1.8x effective magnification and a step-size of 5 µm. Each subject’s microscopic imaging data was co-registered to its respective MRI anatomical dataset and subsequently co-registered to the Allen Brain atlas using ANTs.
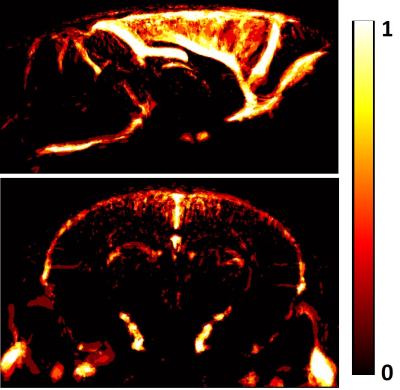
Results: The developed atlas provides a multi-scale representation of the vascular architecture of the mouse brain (Fig.1) and has been co-registered to the Allen brain atlas allowing registration of multimodal information. More specifically, the atlas contains the macro-vasculature acquired with TOF-MRA providing: a) a template of mainly the larger vessels and sinuses (Fig. 1A) and b) a vascular probability map that reflects spatial reliability across subjects with the large vessels presenting very high spatial reliability in contrast to further branches that are more variable (Fig. 2). This also demonstrates that larger vessels should be used for co-registration within and across imaging modalities. Moreover, the atlas also contains the novel double-labelled micro-vasculature. Preliminary results show that the isolectin staining was more effective in labeling the capillaries while albumin staining more successfully labeled the bigger vessels and could be used for alignment to the TOF-MRA. By combining signals of both channels an improved detection of the mouse cerebral vascular architecture can be achieved. Furthermore, the atlas also contains 3D-T2 weighted anatomical information which can be used as an access point for other MRI experiments and to co-register to the Allen brain atlas.
Conclusion: This multi-scale approach provides not only the full vascular tree with unprecedented detail but also a means for alignment and multivariate analysis of multimodal imaging information via the Allen Brain framework. Future work can further extent this atlas with quantitative computational tools able to identify vascular changes and abnormalities as often observed in neurodegenerative disorders.
Acknowledgements
This research was supported by Molecular Imaging of Brain Pathophysiology (BRAINPATH) under grant agreement number 612360 within the Marie Curie Actions-Industry-Academia Partnerships and Pathways (IAPP) program, by the Fund for Scientific Research Flanders (FWO G048917N) and Flagship ERA-NET (FLAG-ERA) FUSIMICE (grant agreement G.0D7651N)References
No reference found.Figures