0368
Unraveling Cardiac and Respiratory Contributions to Brain Tissue Motion using Single Shot 2D DENSE at 7T MRI.1Radiology, University Medical Center Utrecht, Utrecht, Netherlands, 2Neurology, University Medical Center Utrecht, Utrecht, Netherlands
Synopsis
The cardiac cycle and respiration both influence CSF dynamics and therefore the displacement of brain tissue. In this work we unravel their contribution to brain tissue displacement using a single shot 2D cine displacement-encoded imaging method employing stimulated echoes (DENSE) for brain motion measurements. Displacement-encoded data sets in the Feet-to-Head direction of seven volunteers were fitted to a linear model. Consistent trends in displacements were observed. The developed DENSE sequence results showed similar sized contributions to brain tissue displacement. Relating these displacements to contributions to the clearance system remains future work.
Introduction
Brain tissue motion induced by the cardiac and respiration cycles is considered to be involved in the drainage of cerebral waste1. Although cardiac and respiratory pulsations were claimed to play different roles2, this was based on T2* weighted measurement which reflect both blood flow and tissue motion together. We aim to unravel the influence of cardiac and respiratory contributions by specifically measuring the displacement of brain tissue with a single shot 2D DENSE acquisition and fitting a linear model to the observed data.Method
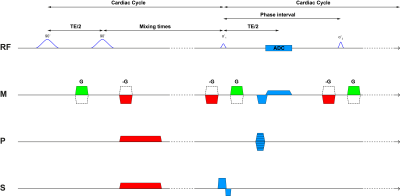
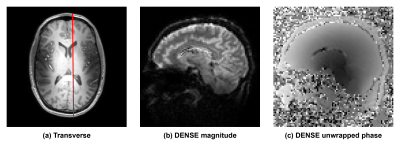
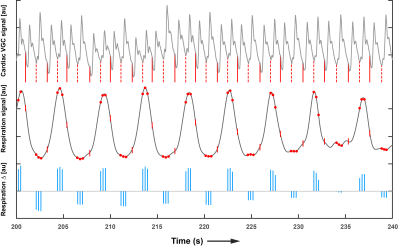
A cardiac triggered, single shot TFEPI 2D cine-DENSE sequence3 was designed to measure heart beat and respiration related brain tissue motion (Figure 1). The displacement sensitivity was set to $$$D_{enc}$$$=0.35 mm/$$$\pi$$$ in the Feet-to-Head direction (resolution 2.98x3.11x3mm3 and FOV 250x250mm2, TFE factor 3, EPI factor 15, SENSE 1.8 (AP direction), TE/2 9ms, and three frames at respectively 0, 23 and 46% of the average cardiac cycle). A variable excitation flip angle $$$\alpha_{n}$$$ over the cardiac cycle was recursively calculated4 using a T1 value of 1100ms and a maximum flip angle of 30˚. Written informed consent was obtained from all volunteers in accordance with the Ethical Review Board of our institution. Seven healthy subjects (3 females, age 25 ± 3 years) were included and scanned at 7T (Philips Healthcare, Best, The Netherlands) using a 32 channel head coil. The 2D cine-DENSE measurements were repeated over 200 dynamics with alternating encoding direction, resulting in 600 snapshots over time, and a scan duration of 5-10 min (80-40bpm). Slice planning is shown in Figure 2. Physiological data was recorded during scanning using a vector cardiogram (VCG) for triggering and a respiration belt to trace abdominal breathing (Figure 3).Analysis
The measured apparent displacement $$$d$$$ of brain tissue at cardiac phase $$$c$$$ and respiration position $$$r$$$, was assumed to be a linear independent combination of cardiac and respiratory displacement contributions represented by $$$d_{c,i}$$$ and $$$d_{r,delta}$$$ respectively. The static phase errors and respiratory induced $$$B_{0}$$$ offsets were described by $$$d_{0}$$$ and $$$d_{r,off}$$$, respectively, yielding the model
$$d(c,r) = \sum_{i=1}^{3} x_{c,i} d_{c,i}+x_{r,delta} d_{r,delta}+x_{r.meas}+d_{r,off}+d_{0}$$
Weighting factors $$$x$$$ were deduced from the physiological data, after smoothing the respiration signal by Fourier filtering between 0.1 and 1 Hz. Depending on the alternating encoding polarity, $$$x_{c,i}$$$ had alternating signs for odd or even dynamic $$$n$$$. For $$$i$$$ is 1, 2 or 3, $$$x_{c,i}=(-1)^{n}$$$ if a frame was acquired at respectively 0, 23 or 46% of the associated actual cardiac interval. Cardiac weighting factors were linearly redistributed for frames that fell between these intervals, such that
$$ 0 \leq \parallel x_{c,i} \parallel \leq 1$$
$$ \sum_{i=1}^{3} \parallel x_{c,i} \parallel = 1 $$
The respiration trace was described in the model by coefficients $$$x_{r,delta}=(-1)^{n} \Delta r$$$ and $$$x_{r,meas} = \Delta r$$$, where $$$\Delta r$$$ is the difference in abdominal respiration position between encoding and decoding. The complete linear model was solved voxelwise by linear regression, using an additional weighting factor proportional to $$$SNR_{\phi} \propto (DENSE_{MAG})^2$$$, where $$$DENSE_{MAG}$$$ is the magnitude of the DENSE image. Wrong triggers were detected and related images were retrospectively discarded for the analysis.
Results and Discussion
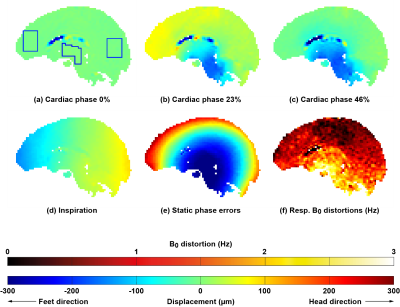
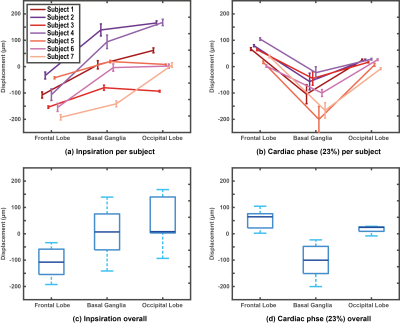
Imaging was successful in all subjects, and triggering was consistent; on average 151 dynamics (range 106 to 199) could be used for analysis. Consistent displacement maps per subject were obtained (Figure 4). Regional analysis was performed in three regions of interest (see Figure 4a for regions) for both cardiac and respiratory motion. Cardiac related displacement was largest in the deep brain (basal ganglia) and was on average -99±30µm and -139±17µm at 23% and 46% of the cardiac cycle, respectively. The results (Figure 5) show that contributions from heart and respiration are in the same order of magnitude. A rotation was observed for the respiratory displacement, where the back of the head moves in the Head direction whereas the front of the head moves in the Feet direction during inspiration and vice versa during expiration. $$$B_{0}$$$ offset fluctuations related to respiration decreased from the base of the brain to the top of the brain, and ranged typically between 0 and 10 Hz, which compares well to the literature5. We did not assess the feasibility of measuring low frequency fluctuations2 (below 0.1 Hz).Conclusion
The developed single shot 2D DENSE sequence is capable of consistently unraveling cardiac and respiratory related brain motion, taking into account offsets due to static phase errors and $$$B_{0}$$$ fluctuations. This method may be useful for MR elastography with intrinsic activation6 and provide insight in the sources of pulsatile brain motion. Relating this motion to contributions to the clearance system remains future work.Acknowledgements
The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement n°337333.References
1. Mestre H, Kostrikov R, Nedergaard M, et al. Perivascular spaces, glymphatic dysfunction, and small vessel disease. Clinical Science. 2017;131(17):2257‐2274.
2. Kiviniemi V, Wang X, Nedergaard M, et al. Ultra-fast magnetic resonance encephalography of physiological brain activity – Glymphatic pulsation mechanisms? Journal of Cerebral Blood Flow & Metabolism. 2016;36(6):1033‐1045.
3. Soellinger M, Rutz M, Boesiger P, et al. 3D cine displacement-encoded MRI of pulsatile brain motion. Magnetic Resonance in Medicine. 2009;61(1):153‐162.
4. Fischer S, McKinnon G, Boesiger P, et al. Improved myocardial tagging contrast. Magnetic Resonance in Medicine. 1993;30(2):191‐200.
5. Van Gelderen P, De Zwart J, Duyn J, et al. Real-time shimming to compensate for respiration-induced B0 fluctuations. Magnetic Resonance in Medicine. 2007;57(2):362‐368.
6. Weaver J, Pattison A, Paulsen K, et al. Brain mechanical property measurement using MRE with intrinsic activation. Physics in Medicine and Biology. 2012;57(22):7275‐7287.
Figures