0256
A Novel Method for Fast and Efficient Measurement of Diffusion Tensor Size and Shape Distributions1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States
Synopsis
We demonstrate through simulations and empirical data that it is possible to simultaneously estimate the variance of the voxel-wise diffusion tensor shape and size distributions using efficient isotropic and linear diffusion encodings on a whole-body clinical MRI scanner with whole-brain coverage at 3mm isotropic resolution in under 2 minutes.
Introduction
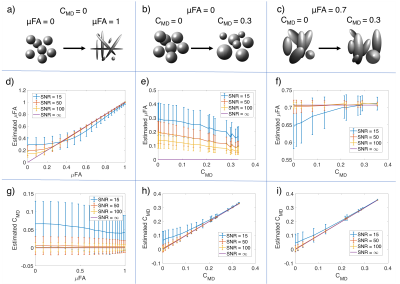
Characterizing the complex microenvironments in the brain is a primary challenge of diffusion MRI. Recent advances in gradient waveform design and modeling have enabled increased specificity to changes in compartment shape and size through advanced diffusion waveforms1-3(Fig. 1a-c) and modeling4-8. The diffusion tensor distribution (DTD) model6 extends the diffusion tensor (DT) model9 by adding a fourth-order tensor($$$\mathbb{C}$$$), describing the covariance of the distribution of DTs. Rotationally invariant measures of size variation (CMD) and microscopic diffusion anisotropy (μFA)(Fig. 1d) are derived from the 21 independent elements of $$$\mathbb{C}$$$. We demonstrate that CMD and μFA can be derived from the orientation averaged $$$\mathbb{C}_{iso}$$$ with fewer measurements than required to fit the full DTD model.Theory
The DTD model approximates the diffusion signal using a fourth-order diffusion covariance tensor:
$$S(\mathbf{B}){\approx}S_0{\exp}(-{\langle}\mathbf{B},\langle{\mathbf{D}\rangle\rangle+\frac{1}{2}\langle}\mathbb{B},\mathbb{C}\rangle)\hspace{3cm}(1)$$
where $$$\mathbf{B}=\int_0^\tau\mathbf{q}(t)\mathbf{q}^\mathrm{T}\,d\tau$$$ is the b-tensor, $$${\langle}\mathbf{D}{\rangle}$$$ is the average DT, $$$\mathbb{B}=\mathbf{B}^{\otimes2}$$$, and $$$\mathbb{C}\in\mathbb{R}^{3\times3\times3\times3}$$$ is the diffusion covariance tensor. Equation 1 can be written in matrix vector form:
$$\left(\begin{array}{c}{\log}S_1\\\vdots \\{\log}S_m\end{array}\right)=\left( \begin{array}{ccc}1&-\mathbf{b}_1^\mathrm{T}&\frac{1}{2}\mathbb{b}_1^\mathrm{T} \\\vdots&\vdots&\vdots\\1& -\mathbf{b}_m^\mathrm{T}&\frac{1}{2}\mathbb{b}_m^\mathrm{T} \end{array}\right)\left(\begin{array}{ccc}\log S_0&\langle\mathbf{d}\rangle&\mathbb{c}\end{array}\right)^\mathrm{T}\hspace{3cm}(2)$$
where $$$\mathbf{b}=\left(\begin{array}{cccccc}b_{xx}&b_{yy} & b_{zz}&\sqrt{2}b_{yz}&\sqrt{2}b_{xz} &\sqrt{2} b_{xy}\end{array}\right)^\mathrm{T}$$$, $$$\mathbb{b}\in\mathbb{R}^{21\times1}$$$ contains the independent elements of $$$\mathbf{b}\mathbf{b}^{\mathrm{T}}$$$, $$$ \mathbf{d}=\left(\begin{array}{cccccc}d_{xx}&d_{yy}&d_{zz}&\sqrt{2}d_{yz}& \sqrt{2}d_{xz}&\sqrt{2}d_{xy}\end{array}\right)^\mathrm{T}$$$ are independent elements of $$$\langle \mathbf{D}\rangle$$$, and $$$\mathbb{c}\in\mathbb{R}^{21\times1}$$$ contains independent elements of $$$\mathbb{C}$$$. However, since CMD and μFA are rotationally invariant, they can be estimated from $$$\mathbb{C}$$$ averaged over all orientations ( $$$\mathbb{C}_{iso}$$$). Since $$$\mathbb{C}_{iso}$$$ describes an isotropic medium, the same signal ($$$S_{iso}$$$) is expected from all diffusion encoding orientations. Therefore $$$\mathbb{b}_{linear}^\mathrm{T}\mathbb{c}_{iso}=\alpha $$$ and $$$\mathbb{b}_{planar}^\mathrm{T}\mathbb{c}_{iso}=\alpha$$$. This can be rewritten as the homogeneous equation: $$$\mathbf{A}\left(\begin{array}{ccc}\mathbb{c}_{iso}&\alpha&\beta\end{array}\right)=0$$$. The null space of $$$\mathbf{A}$$$ can be spanned by two vectors and fully parameterizes $$$\mathbb{c}_{iso}$$$ such that $$$\mathbb{c}_{iso}=\mathbf{V}\mathbf{v}$$$, where $$$\mathbf{V}\in\mathbb{R}^{21\times2}$$$, $$$ \mathbf{v}\in\mathbb{R}^{2\times1}$$$. Therefore, Equation 2 simplifies to:
$$\left(\begin{array}{c}{\log}S_{iso,1}\\\vdots\\{\log}S_{iso,m}\end{array}\right)=\left( \begin{array}{ccc}1&-b_1&\frac{1}{2}b_1^2\mathbf{r}_1\\\vdots&\vdots&\vdots\\1&-b_m&\frac{1}{2}b_m^2\mathbf{r}_m\end{array}\right)\left(\begin{array}{ccc}\log S_0 &\mathrm{MD}&\mathbf{v}\end{array}\right)^\mathrm{T} \hspace{3cm}(3)$$
, where $$$\mathbf{r}_n = \mathbb{b}_n^\mathrm{T}\mathbf{V}/\mathrm{b}_n^2$$$ and $$$S_{iso, n}$$$ can be computed for anisotropic mediums by averaging over multiple orientations. The advantages of Equation 3 over Equation 2 include: 1) it is computationally and numerically advantageous to fit 4 variables compared to 28, 2) estimating $$$S_{iso}$$$ requires fewer encoding directions compared to estimating $$$\mathbb{C}$$$ and 3) both CMD and μFA can be estimated using only linear and isotropic b-tensors, thereby avoiding ellipsoidal or planar b-tensors, which require longer TEs.
Methods
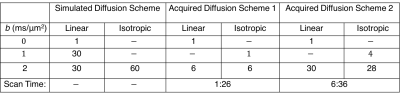
The diffusion environments shown in Figure 3a-c were simulated with the diffusion encoding scheme described in Table 1. Rician noise was introduced to the signal to produce 10,000 noisy measurements with SNRs of 15, 50, and 100 for the non-diffusion weighted signal. Equation 3 was fitted to the noisy signal and the mean and standard deviation of the CMD and μFA were computed6.
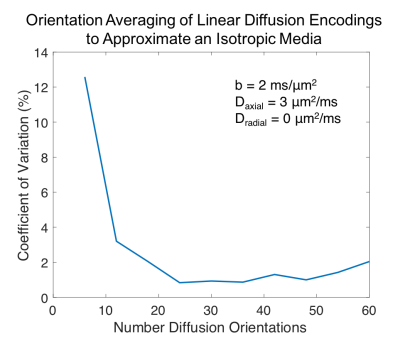
Orientation averaging to approximate $$$S_{iso}$$$ was tested by simulating the worst-case scenario (Daxial=3μm2/ms, Dradial=0μm2/ms). The coefficient of variation(COV) of the diffusion signal across 150 tensor orientations equally spaced on a sphere was computed for diffusion schemes containing 6 to 60 diffusion directions.
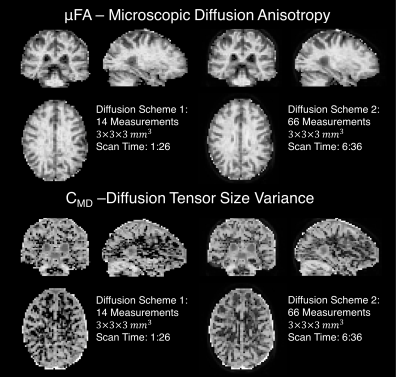
Two healthy subjects were scanned with IRB approval using a 3T whole-body MR system(Premier, GE Healthcare) equipped with a 32-channel head coil(Nova Medical). Each subject was examined with linear and isotropic diffusion encoding sequences(Fig. 1) using diffusion acquisition schemes shown in Table 1. The eddy current and motion corrected10,11 data was fit to Equation 3 to compute CMD and μFA6.
Results
Figure 2 quantifies the rotational invariance achievable using a given number of linear diffusion encodings. Figure 3d-i display μFA and CMD estimates from simulated diffusion signals from linear and isotropic diffusion encodings. Figure 4 shows full brain estimates of μFA and CMD at $$$3\times3\times3$$$mm3 resolution with acquisition times under 2min.
Discussion
Even in the worst-case scenario, a COV under 4% can be achieved with only 12 linear diffusion encodings. This allows estimation of μFA and CMD with 15 (12 linear+B0+isotropic with 2 b-values) measurements compared to the 28 measurements needed to estimate the full DTD. Note that the matrix from Equation 2 is not full rank with any number of linear and isotropic diffusion encodings, and therefore fitting the full DTD is intractable. Both the simulated and in vivo results demonstrate robust estimation of μFA and CMD. The μFA and CMD contrast in the brain is consistent with prior work6, but furthers these efforts by dramatically reducing the scan time and improving brain coverage. Our in vivo results indicate that robust estimates can be achieved with less than 12 linear encodings, and therefore it is advantageous to acquire additional averages of isotropic diffusion encodings to equalize the SNR across linear and isotropic encodings.Conclusion
We demonstrate the simultaneous estimation of compartment shape and size dispersion using efficient isotropic and linear diffusion encodings.Acknowledgements
Funding provided by an NSF-GRFP, NIH: R01-NS095985, S10-RR026351, P41-EB015891, GE Healthcare and the Dana Foundation.References
1. Sjolund J, Szczepankiewicz F, Nilsson M, Topgaard D, Westin CF, Knutsson H. Constrained optimization of gradient waveforms for generalized diffusion encoding. J Magn Reson 2015;261:157-68.
2. Mitra PP. Multiple wave-vector extensions of the NMR pulsed-field-gradient spin-echo diffusion measurement. Phys Rev B Condens Matter 1995;51(21):15074-15078.
3. Eriksson S, Lasic S, Topgaard D. Isotropic diffusion weighting in PGSE NMR by magic-angle spinning of the q-vector. J Magn Reson 2013;226:13-8.
4. Jespersen SN, Lundell H, Sonderby CK, Dyrby TB. Orientationally invariant metrics of apparent compartment eccentricity from double pulsed field gradient diffusion experiments. NMR Biomed 2013;26(12):1647-62.
5. Lawrenz M, Koch MA, Finsterbusch J. A tensor model and measures of microscopic anisotropy for double-wave-vector diffusion-weighting experiments with long mixing times. J Magn Reson 2010;202(1):43-56.
6. Westin CF, Knutsson H, Pasternak O, Szczepankiewicz F, Ozarslan E, van Westen D, Mattisson C, Bogren M, O'Donnell LJ, Kubicki M and others. Q-space trajectory imaging for multidimensional diffusion MRI of the human brain. Neuroimage 2016;135:345-62.
7. Szczepankiewicz F, Lasic S, van Westen D, Sundgren PC, Englund E, Westin CF, Stahlberg F, Latt J, Topgaard D, Nilsson M. Quantification of microscopic diffusion anisotropy disentangles effects of orientation dispersion from microstructure: applications in healthy volunteers and in brain tumors. Neuroimage 2015;104:241-52.
8. Lasič S, Szczepankiewicz F, Eriksson S, Nilsson M, Topgaard D. Microanisotropy imaging: quantification of microscopic diffusion anisotropy and orientational order parameter by diffusion MRI with magic-angle spinning of the q-vector. Frontiers in Physics 2014;2:11.
9. Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Di Chiro G. Diffusion tensor MR imaging of the human brain. Radiology 1996;201(3):637-48.
10. Andersson JL, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 2016;125:1063-78.
11. Andersson JL, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 2003;20(2):870-88.
12. Yang G, Tian Q, Leuze C, Wintermark M, McNab J. Visualizing Axonal Damage in Multiple Sclerosis Using Double Diffusion Encoding MRI in a Clinical Setting. Proc. Intl. Soc. Mag. Reson. Med. 25 (2017) 2017.
Figures
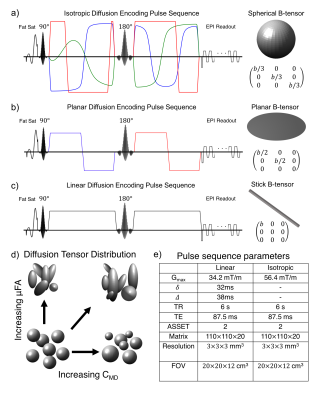
Figure 1: Q-space trajectory imaging employs time varying diffusion waveforms to produce isotropic (a) planar (b) and linear (c) diffusion encoding as described by the appearance of the resulting b-tensor. Red, green, and blue gradient waveforms represent orthogonal gradient axes. By varying the b-tensor shape, information can be obtained about the diffusion tensor distribution (d), which is inaccessible using only conventional linear diffusion encoding. The diffusion tensor distribution can be used to disentangle microstructural shape and size distributions (d). Pulse sequence parameters for the linear and isotropic diffusion encoding pulse sequences used in this study (e).