0255
Unconstrained Estimation of Microstructure by the Combination of Single- and Double-planar diffusion encoding1University Freiburg, Faculty of Medicine, Freiburg, Germany
Synopsis
Modeling of white matter microstructure using standard diffusion MRI protocols is a poor-conditioned problem. With standard measurements there is a global degeneracy: There are two significantly different microstructural parameter sets explaining experimental data equally well. In this work we use an additional planar double diffusion encoding to resolve the degeneracy, and extend a Bayesian estimation framework to a combination of linear and planar diffusion encoding. This enables us to produce reliable estimates without applying any constraints to the microstructural diffusion model.
Introduction
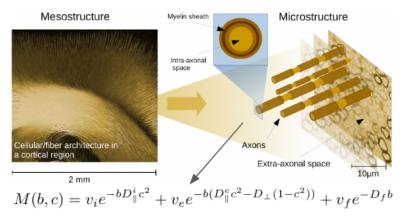
Modeling of the white matter microstructure using diffusion MRI is a poor-conditioned problem, because mesostructure, the fiber architecture at the subvoxel scale, is a strong confounding factor. Recently, it became apparent that, although micro- and mesostructural information is deeply interwoven, it can be estimated fairly independently. Low order orientation features are nearly sufficient to fit microstructural parameters (1,2,3) and even zero-order features (the spherical means) are already quite informative. However, the parameter determination with the common measurement techniques suffers from a global degeneracy: There are two significantly different microstructural parameter sets explaining experimental data equally well (4). Unambiguous parameter determination is only possible using additional data; in the absence of such data, additional constraints or assumptions are necessary. In our companion abstract (6108) we have shown in simulations that an additional planar double diffusion encoding PDE (6,7) can resolve the above mentioned degeneracy. With this knowledge, we extended the Bayesian framework proposed in (2) to a combination of linear encoding (SDE) and planar encoding (PDE) measurements. We show that microstucture imaging with this combination is able to produce reliable estimates without applying any constraints to the prior distributions.Method
We acquired a 4x4x4mm EPI, TE=140ms, TR=3000ms, pF 6/8 with three directional shells (b=100,1000,2000 $$$\text{s/mm}^2$$$) and four planar shells (b=500,1500,3500,5000 $$$\text{s/mm}^2$$$) for three subjects. Since a planar encoding is characterized by the direction of the normal to the encoding plane, the PDE can be treated on the same footing as the SDE. Furthermore, the multiplicative behaviour in the spherical harmonic domain of meso- and micro-structural representations is also preserved. Therefore, conceptually there is no difference between ‘planar’ and ‘directional’ shells - they can be simply merged and integrated into the existing framework (2). Following the standard model (see Figure 1) and (2), we used features up to spherical order 2 while $$$b_0$$$ information was disregarded (we use only ratios of rotation invariants). So we have overall $$$N = 2(n_\text{shells}-1) = 2(7-1)=12$$$ meso invariant features to estimate five micro-structural parameters. A polynomial of order three was used to map these N features onto the microstructural parameters. The prior distributions were the same as in (2), but without any constraints for resolving the bimodality.Results
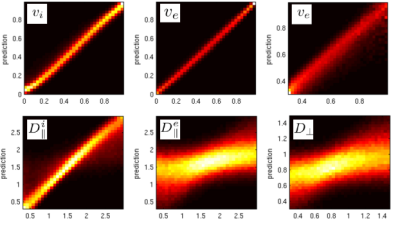
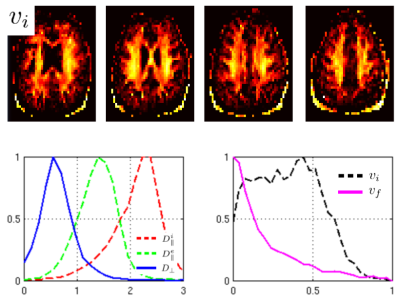
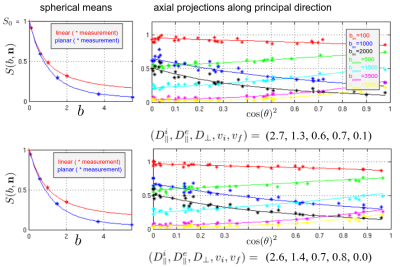
In Figure 2, we show correlations of the trained polynomial regressor with the ground truth. Figure 3 shows maps for some selected slices of the intra axonal signal fraction and histograms of the parameters in white matter. Figure 4 shows signal courses of two example voxels taken from the posterior limb of the internal capsule (PLIC): Spherical averages together with the predicted signal courses against $$$\cos(\theta)^2$$$, where $$$\theta$$$ is the angle to the principal direction, are shown.Discussion
The present unconstrained method demonstrates a better correlation with the ground truth than the single direction “only” setting (2), in which an explicit branch selection was made. Only the extra-axonal diffusivities still show a significant uncertainty due to the shallow optimization landscape. Training exclusively on the spherical averages shows only a slightly worse quality (not shown). Looking at the distribution of parameters, we find means of $$$(D^i_{||},D^e_{||},D_\perp) = (2.25, 1.45, 0.55) \mu m^2/ms $$$ for the diffusivities, which are close to what is reported elsewhere. However, specifically in areas with high intra-axonal fraction our estimates of $$$D^i_{||}$$$ are consistently higher than what was reported (at around 2.7). Two examples from the PLIC are shown in Figure 3. The fits are good and have normalized log likelihoods that are close to one (1.05 and 0.98) at an estimated SNR level of 55. High values of $$$D^i_{||}$$$ remain to be understood in terms of intra-axonal structure. The mean estimator of $$$D^i_{||}$$$ actually tends to underestimate it for high values (see Fig 2), which makes it more likely to believe in the prediction. Since, the mesostructural averaging destroys lots of microstructural information, we can estimate only the mean of the parallel diffusivities in a faithful way. Overall, the presented approach offers the possibility to estimate microstructural diffusion parameters in an fully unconstrained way. We presented a machine-learning based analysis, however also classic fitting/MLE-based approaches (1,2,7) highly benefit from the combination of SDE and PDE measurements. The exact values of the diffusivities still remain uncertain due to the shallow likelihood. Values may be improved by additional measurements (e.g. TripleDE, (5) ) or higher resolution (less mesostructural averaging), in this study a very coarse resolution was used for the sake of SNR.Acknowledgements
No acknowledgement found.References
(1) Kaden, Enrico, et al. "Multi-compartment microscopic diffusion imaging." NeuroImage 139 (2016): 346-359.
(2) Reisert, Marco, et al. "Disentangling micro from mesostructure by diffusion MRI: A Bayesian approach." NeuroImage 147 (2017): 964-975.
(3) Novikov, Dmitry S., et al. "Mapping orientational and microstructural metrics of neuronal integrity with in vivo diffusion MRI." arXiv preprint arXiv:1609.09144 (2016).
(4) Jelescu, Ileana O., et al. "Degeneracy in model parameter estimation for multi‐compartmental diffusion in neuronal tissue." NMR in Biomedicine 29.1 (2016): 33-47.
(5) Dhital, Bibek, et al. "The Absence of Restricted Water Pool in Brain White Matter", NeuroImage accepted, (2017)
(6) Westin, Carl-Fredrik, et al. "Q-space trajectory imaging for multidimensional diffusion MRI of the human brain." Neuroimage 135 (2016): 345-362.
(7) Coelho, Santiago, et al. "Double Diffusion Encoding vs Single Diffusion Encoding in Parameter Estimation of Biophysical Models in Diffusion-Weighted MRI", ISMRM 2017
Figures