0246
3D Dynamic Hyperpolarized-13C Parallel MRI of Human Brain using SVD Low-Rank Matrix Completion1Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States, 2Department of Radiology, Chonnam National University Medical School and Hospital, Gwangju, Republic of Korea
Synopsis
The goal of this feasibility study was to develop and apply new 3D dynamic hyperpolarized 13C combined parallel imaging + compressed sensing MRI methods for human brain studies. The new framework utilizing multichannel coils and low-rank matrix completion reconstruction provided greatly improved coverage as compared to prior 2D acquisitions. Whole-brain coverage was achieved, while maintaining the image quality in terms of spatial distribution, temporal dynamics and quantitative accuracy of the 13C biomarkers. Animal studies and simulations also showed that the pMRI+CS framework provided improved performance over CS alone, and was able better to recover low-SNR peaks.
Purpose
Hyperpolarized 13C MRI studies of human brain1, pelvis and abdomen2 can benefit from array coils to achieve larger coverage and higher sensitivity, and accelerated acquisition techniques for the limited HP T1 relaxation time. Historically, parallel imaging (pMRI) and compressed sensing (CS) were considered independent techniques. Recent developments in the theory of low-rank matrix completion unify the two by first consolidating the sparsity and coil-domain redundancy into a framework of structured-Hankel matrix, and then apply low-rankness for k-space interpolation3,4,5. In this study we conducted first ever HP-13C 3D dynamic parallel+CS MR studies of human brain using an 8-channel receiver array and a randomly-undersampled EPSI acquisition, and applied a SVD-based low-rank matrix completion reconstruction3,4. Performance of the new framework was evaluated using preclinical rat studies and simulations.Methods
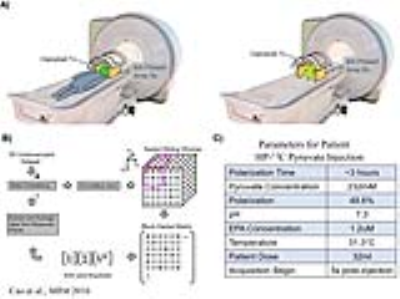
Sequences: A 3D dynamic CS-EPSI sequence was developed to acquire undersampled human brain and rat datasets6,7. The parameters for human brain exam was: spatial resolution=2x2x2cm, temporal=3s, FOV=24x24x32cm, matrix size=12x12x16x59(spectral points)x24(timepoints)x8(channels); for rat: spatial resolution=8x8x8mm, temporal=2s, FOV=9.6x9.6x12.8cm, matrix size=16x16x16x59x21x8. A constant 10° flip angle was applied for the human brain, while for rat studies variable flip angles for each metabolite were used8.
MRI experiments: A clamshell volume transmitter and 8-channel receive array were used for this study (Fig 1A). For the brain patient exam, GMP-grade [1-13C]pyruvate was polarized in a 5T SPINlab polarizer. Parameters of patient HP-pyruvate injection were summarized in the table in Fig.1C. The 2 healthy Sprague Dawley rats received 3ml of 80mM pyruvate bolus (3.35T SPINlab, 25-35% polarization) over 12 seconds, and sequence started 3s post-injection. All studies were approved by IRB and IACUC.
Simulations: Simulation data were constructed using coil sensitivity map acquired from phantom. Temporal dynamics was generated using a two-site model9 (kPL = 0.04s-1 and T1 = 30s based on reported in vivo values). Gaussian noise of 10-45dB peak SNR was added to the dataset, and the data was undersampled using the same pseudorandom pattern as human brain study.
Reconstruction Pipeline: A block Hankel matrix was constructed using 3D sliding window along kx, ky and kf dimensions of the 5D undersampled dataset (Fig.1B). Dynamic and coil dimensions were stacked along row and column directions, respectively. Low-rankness was applied on the block Hankel matrix using singular value decomposition (SVD) and soft thresholding, followed by enforcement of data consistency. This used an iterative process that interpolated the missing k-space points. The reconstruction pipeline was implemented on a high-performance computing (HPC) platform for efficiency. Conventional channel-by-channel CS reconstruction was also conducted as comparison.
Results and Discussions
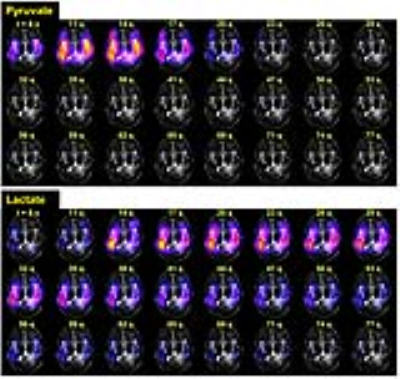
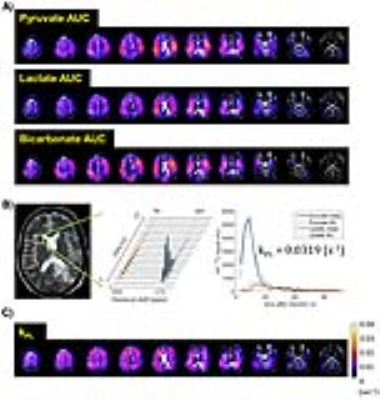
The dynamics in this GBM patient showed HP-13C pyruvate signal appearing about 8 seconds after the end of injection (Fig.2). The product lactate was observed at around the same time, reflecting the rapid pyruvate-lactate conversion in human brain. The lactate reached maximum around 20 seconds, and lasted until 68-71 seconds post-injection. The spatial distributions of HP-13C pyruvate, lactate and bicarbonate were illustrated in Fig.3A. Fig.3C shows the kPL map, where estimated kPL=0.0319s-1 was found in a well-perfused voxel of normal brain (Fig.3B). These findings were highly consistent with the prior 2D human brain data1. It indicated that the 3D-EPSI+SVD reconstruction provided greater coverage of the whole brain, while maintaining the quality of prior 2D EPSI acquisitions.
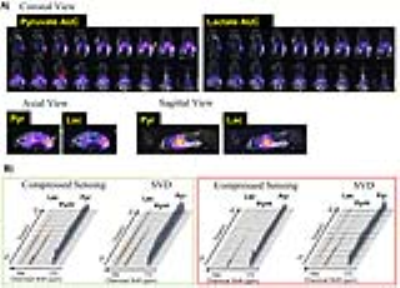
Figure 4A illustrates the distribution of pyruvate and lactate in a rat. As shown in Fig.4B, in a high-signal kidney voxel (green box), mean SNR was 177 and 78 for pyruvate and lactate using SVD, compared to 139 and 65 using individual-channel CS. In a liver voxel (red box) that had relative low SNR, SVD provided mean SNR of 22.8 and 6 for pyruvate and lactate, whereas CS provided 12.4 and 2.5. Note lactate signal was barely detectable in the CS reconstructed spectrum, and the difference was conspicuous after ~35 seconds, where only SVD recovered the latter half of lactate dynamics.
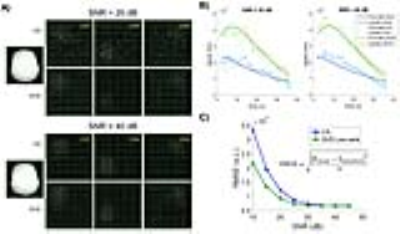
Simulation also demonstrated that, under low SNR conditions, SVD reconstruction produced less “speckle” like noise compared to CS (Fig.5A), had lower bias (Fig.5B), and lower root mean-squared error (Fig.5C).
In summary, the SVD-based low-rank matrix completion outperformed CS technique alone by simultaneously taking advantage of coil-domain redundancy (pMRI) and data sparsity (CS).
Conclusions
This study showed the feasibility of conducting parallel+CS hyperpolarized 13C MRSI on human brain at high speed and with whole-brain coverage, enabled by an undersampled multichannel acquisition in tandem with a reconstruction strategy based on the theory of low-rank matrix completion. This novel framework has great potential in future studies of metabolic reprogramming in human brain malignancies and other neurological diseasesAcknowledgements
This work was supported by grants from the NIH (R01EB017449, R01EB013427, R01CA166655, and P41EB013598).References
[1] Park I et al., Dynamic Hyperpolarized 13C Metabolic Imaging of Patients with Brain Tumors. Proceedings of the 25th Annual Meetings of ISMRM 2017;
[2] Zhu Z et al., Hyperpolarized 13C Dynamic Breath-held Molecular Imaging to Detect Targeted Therapy Response in Patients with Liver Metastases. Proceedings of the 25th Annual Meetings of ISMRM 2017;
[3] Shin PJ et al., Calibrationless Parallel Imaging Reconstruction Based on Structured Low‐rank Matrix Completion, Magn. Reson. Med. 2014; 72(4): 959-970.
[4] Cao P et al., Accelerated High‐bandwidth MR Spectroscopic Imaging Using Compressed Sensing, Magn. Reson. Med. 2016; 76(2): 369-379.
[5] Jin KH et al., A General Framework for Compressed Sensing and Parallel MRI Using Annihilating Filter Based Low-Rank Hankel Matrix. IEEE Trans Comput Imaging. 2016; 2(4), 480-495.
[6] Hu S et al., 3D Compressed Sensing for Highly Accelerated Hyperpolarized C-13 MRSI With In Vivo Applications to Transgenic Mouse Models of Cancer. Magn. Reson. Med. 2010; 63(2):312-21.
[7] Larson PEZ et al., Fast Dynamic 3D MR Spectroscopic Imaging with Compressed Sensing and Multiband Excitation Pulses for Hyperpolarized C-13 Studies. Magn. Reson. Med. 2011; 65(3):610-9.
[8] Xing Y et al., Optimal variable flip angle schemes for dynamic acquisition of exchanging hyperpolarized substrates. J. Magn. Reson. 2013; 234, 75-81.
[9] Bahrami N et al., Kinetic and perfusion modeling of hyperpolarized 13C pyruvate and urea in cancer with arbitrary RF flip angles. Quant Imaging Med Surg. 2014; 4(1):24
Figures