0222
Iron-related gene expression associated with magnetic susceptibility reductions: Application to the pathophysiology of a movement disorder population1Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 2Department of Psychiatry, Hannover Medical School, Hannover, Germany, 3Siemens Healthcare, Erlangen, Germany, 4Douglas Mental Health Institute, McGill University, Montreal, QC, Canada, 5Massachusetts General Hospital, Harvard Medical School, Charlestown, MA, United States
Synopsis
We employ a genetic-imaging approach to examine the underlying genetic basis of magnetic susceptibility reductions at a major locus of pathophysiology in Gilles de la Tourette syndrome (GTS). Voxel-wise statistical differences of motor-striatal susceptibility exhibited significant associations with the expression profile of iron-related gene-sets extracted from the Allen Human Brain Atlas, thus suggesting that the expression of iron-related genes coincides with patterns of susceptibility reductions in GTS. This work supports previous studies relating magnetic susceptibility to brain iron and provides an example of an analytic strategy in which valuable insights can be gleaned by exploring associations between gene-expression and image-derived phenotypes.
Introduction
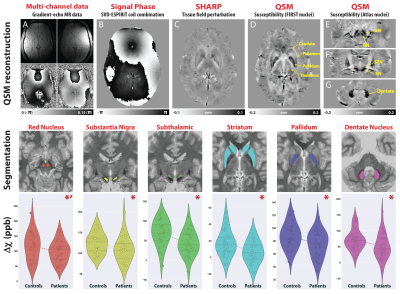
Gilles de la Tourette syndrome (GTS) is a neuropsychiatric movement disorder characterized by tics with reported abnormalities in the neurotransmission of dopamine, GABA and glutamate (1, 2). Given that iron plays an integral role in varied biochemical processes involved in neurotransmitter synthesis and transport (3), we hypothesized that iron exhibits a role in GTS pathophysiology. Utilizing Quantitative Susceptibility Mapping (QSM) as a surrogate measure of iron, we showed that GTS patients exhibit magnetic susceptibility reductions in subcortical regions implicated in disease pathophysiology (Fig. 1) (4). To explore the underlying genetic basis of these reductions, we employed an imaging-genetic approach to assess relationships between voxel-wise, nucleus specific, susceptibility differences with the default expression profile of iron-related gene-sets extracted from the Allen Human Brain Atlas (AHBA) (5). Given that genetic transcriptional profiles are known to exhibit distinct expression patterns in the brain, we aimed to investigate spatially specific relationships exhibited between susceptibility and gene expression patterns to glean further insights into pathophysiological mechanisms of iron-related changes. To explore the relevance of these reductions to the clinical population, we additionally employed a machine learning approach to investigate associations with clinical symptomatology.Methods
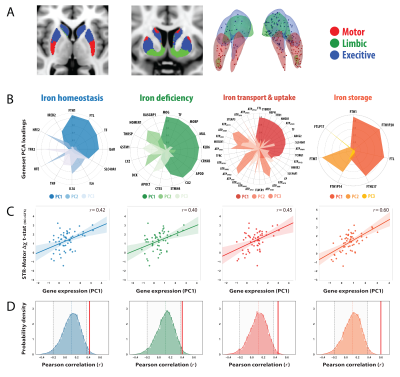
QSM and MP2RAGE data were acquired from 28 GTS patients and 26 age/gender matched healthy controls on a 3T Siemens MAGNETOM Verio using a 32-channel head coil. A 10ml blood sample was collected from each subject for the quantitation of serum Ferritin, in addition to a comprehensive clinical assessment battery. Susceptibility-weighted data were acquired using FLASH (TR=30ms; TE=17ms; flip-angle=13°; 0.8mm isotropic nominal resolution). High-quality phase maps were reconstructed using data-driven coil combination (6) and QSM images were computed using the SDI approach (7) with referencing to lateral ventricle CSF (8). To investigate associations between magnetic susceptibility and clinical symptoms, we first decomposed the clinical data into a set of clinical scores using Principal Component Analysis (PCA). To interrogate genetic mechanisms that may drive abnormalities in iron levels, we employed a cross-correlation approach to examine the relationship between susceptibility differences with gene expression profiles extracted from the AHBA. Voxel-wise susceptibility difference statistical maps were calculated via nonparametric permutation testing while accounting for age, gender and image quality (FSL-randomise, 10,000 permutations). Transcriptional levels of genes incorporated within four iron-related gene sets (iron-homeostasis, iron-deficiency; iron transport and uptake and Iron-storage (9, 10)) were then extracted from loci of pathophysiology at specific coordinates sampled in the AHBA. For each gene-set, principal components were extracted and cross-correlated to statistical values of striatal susceptibility differences in same coordinate space (MNI). To evaluate whether the observed correlations were significant, a permutation-based approach was implemented in which the null distribution was constructed using a re-sampling based approach (10,000 permutations). For each permutation, Pearson correlation was calculated between magnetic susceptibility differences and the average gene-expression value of random set of genes with an equal size to the gene set of interest (alpha level of 0.05).Results
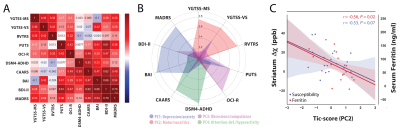
The correlation matrix between all the acquired clinical variables revealed sufficient complementary for data-reduction using PCA (Fig 2A). PCA yielded a set 4 components explaining 77% of the variance that were interpreted as representing scores for (i) depression/anxiety; (ii) motor-tics, (iii) obsessions/compulsions, (iv) attention-deficits/hyperactivity (Fig. 2B). Regression analysis between the motor-tic score and surrogate measures of iron revealed a trend with serum ferritin levels and a significant negative association with striatal susceptibility (Fig. 2C). Driven by these results, iron-related gene expression profiles were extracted within three functionally distinct sub-territories of the striatum (motor, associative, limbic) and cross-correlated with statistical maps of magnetic susceptibility reductions at the same coordinates. Permutation based inference revealed significant positive associations between striatal-motor susceptibility and the principal components of the iron-related gene-sets (Fig. 3). Inspection of associations between the mean expression profile of the iron-related gene-sets and magnetic susceptibility statistical values, revealed similar findings. These results indicate that iron-related abnormalities in the motor sub-division of the striatum exhibit a major role in the pathophysiology of GTS.Discussion
We demonstrate a link between magnetic susceptibility reductions and default expression profiles of iron-related genes within a major locus of pathophysiology in GTS. These findings suggest that the expression profiles of iron-related genes coincide with patterns of susceptibility reductions in patients with GTS, thus providing a link between disrupted iron homeostasis and GTS pathophysiology. This work supports previous studies relating magnetic susceptibility to brain iron and provides an example of an analytic strategy in which valuable insights on disease pathophysiology can be achieved by exploring associations between genetic transcriptional profiles and image derived phenotypes.Acknowledgements
This work was funded by the FP7 Marie Curie Actions of the European Commission (“TS-EUROTRAIN FP7-PEOPLE-2012-ITN, Grant No. 316978”) and, in part, by the Helmholtz Alliance “ICEMED: Imaging and Curing EnvironmentalMetabolic Diseases”.References
- Kanaan AS, Gerasch S, García-García I, Lampe L, Pampel A, Anwander A, et al. (2017): Pathological glutamatergic neurotransmission in Gilles de la Tourette syndrome. BRAIN. 140: 218–234.
- Singer H (2013): The Neurochemistry of Tourette Syndrome. In: Martino D, Leckman JF, editors. Tourette Syndr. Oxford University Press, pp 276–297.
- Bianco L, Unger E, Beard J (2010): Iron Deficiency and Neuropharmacology. In: Yehuda S, Mostofsky DI, editors. Iron Defic Overload From Basic Biol to Clin Med. Totowa, NJ: Humana Press, pp 141–158.
- Kanaan AS, Anwander A, Schäfer A, Bilgic B, Schlumm T, Near J, et al. (2017): QSM meets MRS: The influence of subcortical iron on glutamatergic neurotransmission in a movement disorder population. Proc 25th Annu Meet ISMRM. p 4649.
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. (2012): An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 489: 391–399.
- Bilgic B, Polimeni JR, Wald LL, Setsompop K, States U (2016): Automated tissue phase and QSM estimation from multichannel data. Proc Intl Soc Mag Reson Med 24. Singapore, Singapore. doi: 10.1016/j.neuroimage.2012.05.067.4.
- Schweser F, Deistung A, Sommer K, Reichenbach JR (2013): Toward online reconstruction of quantitative susceptibility maps: Superfast dipole inversion. Magn Reson Med. 69: 1582–1594.
- Straub S, Schneider TM, Emmerich J, Freitag MT, Ziener CH, Schlemmer HP, et al. (2016): Suitable reference tissues for quantitative susceptibility mapping of the brain. Magn Reson Med. doi: 10.10. doi: 10.1002/mrm.26369.
- Clardy S, Wang X, Zhao W, Liu W, Chase G, Beard J, et al. (2006): Acute and chronic effects of developmental iron deficiency on mRNA expression patterns in the brain. J Neural Transm. 71: 173–96.
- Hower V, Mendes P, Torti FM, Laubenbacher R, Akman S, Torti S V (2009): A general map of iron metabolism and tissue-specific subnetworks w. 422–443.
Figures