0218
TOPPE: A framework for rapid prototyping of MR pulse sequences1Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
Synopsis
We present a simple MR pulse programming framework for rapid prototyping of complex, custom MR pulse sequences. We define a file format that contains all low-level details of an MR imaging experiment, along with a driver (or interpreter) that loads the file(s) and executes the experiment on a General Electric scanner. We demonstrate that a wide range of sequences can be implemented with this setup, subject to limitations such as non-overlapping waveforms. Our framework is conceptually similar to Pulseq, a recently-introduced file format specification for which drivers for other vendor platforms exist (at present, Siemens and Bruker).
INTRODUCTION
Implementing novel MR sequences on clinical scanners requires low-level, platform-dependent programming, which is technically challenging and discourages cross-platform collaboration and sequence validation. Recently, several alternative pulse programming platforms have been proposed that ease the task of designing, simulating, and implementing custom sequences, and porting sequences across vendor platforms1-5. Critical to several of these efforts are vendor-specific “drivers”, or “interpreters”, that load a detailed pulse sequence specification from file and executes it on the scanner. Such drivers must be “universal”, in the sense that they should be able to execute arbitrary sequences of gradient/RF waveforms and data acquisition windows. Here we introduce a universal driver for GE scanners, along with a file format specification for describing all low-level pulse sequence details. We also provide file converters between our custom format, which we call “TOPPE”, and Pulseq5, a recently introduced sequence specification compatible with two other vendor platforms (Siemens and Bruker).METHODS
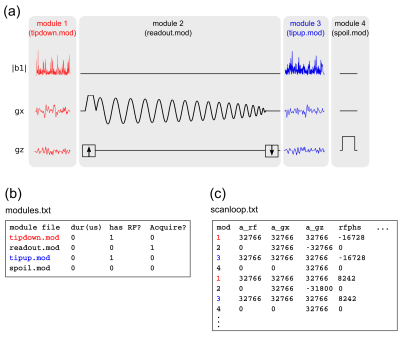
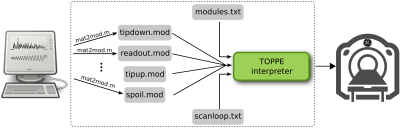
Figure 1 illustrates the three types of files required by TOPPE: (i) two or more ’.mod’ files, (ii) one ’modules.txt’ file, and (iii) one ’scanloop.txt’ file. Each .mod file specifies one unique pulse sequence segment, or module, contained in the planned MR experiment. In the example in Fig. 1, there are four unique modules: two different multi-dimensional RF excitations, one spiral-in readout with kz-encoding, and one gradient spoiler. The .mod files are the basic “building blocks” of the MR experiment, and can be shared (re-used) among different TOPPE sequences. The file modules.txt lists each .mod file contained in the MR experiment. In addition, modules.txt specifies module duration, and whether a module is an RF excitation, data acquisition, or gradients-only module. In this way, RF waveforms or data acquisition windows can be easily turned on or off, which is useful for, e.g., testing or gradient calibration purposes. Finally, scanloop.txt lists the order in which to execute the modules, and specifies waveform amplitudes and other dynamic information (see Fig. 1 caption for details). The data storage location (for rows containing a data acquisition module) is specified using three indices: ’slice’, ’echo’, and ’view’. These indices directly reflect how GE scanners store data internally. The sequence designer is free to use these indices in any manner he or she finds useful, i.e., the data need not adhere to a conventional slice/echo/view structure. Once all TOPPE files have been created, they are loaded by the TOPPE driver (a binary executable) that executes the sequence on the scanner (Fig. 1). The driver itself is universal in the sense that its source code is independent of the specifics of any one sequence; the driver merely loads the modules listed in modules.txt and executes them according to the instructions in scanloop.txt. Thus, the sequence specification (encapsulated in the TOPPE files) is separated from sequence implementation (the driver). Figure 2 shows the overall workflow.RESULTS
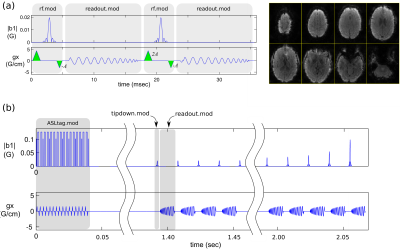
We have successfully implemented and executed a range of sequences; two examples are shown in Fig. 3. By simply changing the various TOPPE sequence files, and without even re-prescribing the sequence (i.e., without changing sequence parameters using the MR console user interface), a single binary executable (driver) was used to execute several different sequences. No sequence-specific compilation is necessary; the driver only needs to be compiled once (for every scanner software upgrade). Open-source Matlab scripts for creating .mod files are available on Github at https://toppemri.github.io/, along with complete sequence examples. Also provided are the Matlab scripts ‘ge2seq.m’ and ‘seq2ge.m’ for converting to and from the Pulseq file format, respectively. EPIC source code for the TOPPE GE driver is also on Github, available to sites with a GE research license (contact the authors for access).CONCLUSION
The TOPPE file format is a complete specification of an MR imaging experiment, based on arbitrary sequences of a (typically small) number of unique modules. Along with the GE driver, TOPPE comprises a modular and flexible platform for rapid prototyping of new pulse sequences. TOPPE is compatible with Pulseq (via a file conversion), a similar platform-independent sequence specification for which drivers for other vendors exist (at present, Siemens and Bruker). The ability to specify a complete MR acquisition in a (mostly) vendor-independent manner and execute that sequence on multiple MR vendor platforms, represents an important step toward truly platform-independent pulse programming.Acknowledgements
This work was supported by NIH grants R01EB023618 and R21EB019653.References
1. Skare S, Avventi E, Norbeck O, Ryden H. An abstraction layer for simpler EPIC pulse programming on GE MR systems in a clinical environment. In Proc., ISMRM, 25th Annual Meeting. Honolulu, 2017; 3813.
2. Magland JF, Li C, Langham MC, Wehrli FW. Pulse sequence programming in a dynamic visual environment: SequenceTree. Magn Reson Med 2016;75:257–265.
3. Debbins J, Gould K, Halleppanavar V, Polzin J, Radick M, Sat G, Thomas D, Trevino S, Haworth R. Novel software architecture for rapid development of magnetic resonance applications. Concepts Magn Reson 2002;15:216–237.
4. Honroth T, Cordes C, Archipovas S, von Samson-Himmelstjerna F, Günther M, Porter D. Advanced platform-independent MR prototyping: From EPI to Arterial Spin Labeling without code compilation. In Proc., ISMRM, 25th Annual Meeting. Honolulu, 2017; 3824.
5. Layton KJ, Kroboth S, Jia F, Littin S, Yu H, Leupold J, Nielsen JF, Stöcker T, Zaitsev M. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn Reson Med 2017; 77:1544–1552.
Figures