0201
Automated Knee Cartilage Segmentation with Very Limited Training Data: Combining Convolutional Neural Networks with Transfer Learning1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States, 3LVIS Corporation, Palo Alto, CA, United States, 4Mechanical Engineering, Stanford University, Stanford, CA, United States, 5Bioengineering, Stanford University, Stanford, CA, United States, 6Neurology & Neurological Sciences, Stanford University, Stanford, CA, United States, 7Neurosurgery, Stanford University, Stanford, CA, United States, 8Orthopaedic Surgery, Stanford University, Stanford, CA, United States
Synopsis
Magnetic resonance imaging is commonly used to study osteoarthritis. In most cases, manual cartilage segmentation is required. Recent advances in deep-learning methods have shown promise for automating cartilage segmentation, but they rely on the availability of large training datasets that rarely represent the exact nature or extent of data practically available in routine research studies. The goal of this study was to automate cartilage segmentation in studies with very few training datasets available by creating baseline segmentation knowledge from larger training datasets, followed by creating transfer learning models to adapt this knowledge to the limited datasets utilized in typical study.
Introduction
Cartilage degradation is one of the hallmarks of osteoarthritis (OA), a leading cause of disability globally1. MRI is commonly utilized in OA research studies to quantify morphological cartilage degeneration2 and to measure T2 relaxation times. Both may be potential biomarkers of OA activity3,4, and both require segmentation of knee cartilage from hundreds of imaging volumes. Standard segmentation techniques are extremely tedious, time-consuming, and costly as they rely on highly-trained readers to manually delineate the cartilage in MR images. Deep-learning methods have shown promise for automating cartilage segmentation5,6,7, but these rely on the availability of large training datasets that are often not available in routine research studies. Thus, the goal of this study was to automate cartilage segmentation in studies with very few training datasets available by creating baseline segmentation knowledge from larger training datasets, followed by using transfer learning to adapt this knowledge to the limited datasets.Methods
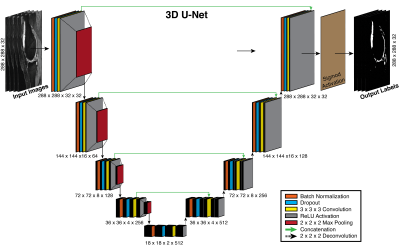
We designed a neural network with a 3D U-Net system architecture (Figure 1), inspired by 2D U-Nets8. In the 3D U-Net, contracting and expanding layers are connected to provide high-resolution contextual features to the deep-layers of the network while minimizing memory usage. These connections better localize high-resolution features and additionally allow deeper networks due to efficient use of memory. Moreover, 3D networks may allow more robust segmentations than 2D by accounting for additional spatial cues.
To create the ‘baseline knowledge’, the 3D U-Net was trained for 50 epochs to segment the femoral cartilage from 3D sagittal double-echo in steady-state (DESS) knee datasets obtained through the Osteoarthritis Initiative (OAI) (relevant parameters: Siemens 3.0T MRI scanners, 8-channel rigid coil, matrix=384x307, 160 slices, slice-thickness=0.7mm, FOV=14cm)9. 124 patients were used for training, 35 for validation, and 17 for testing.
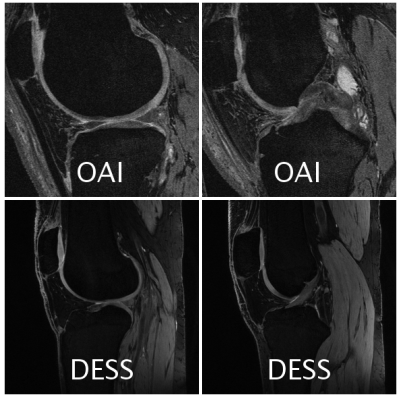
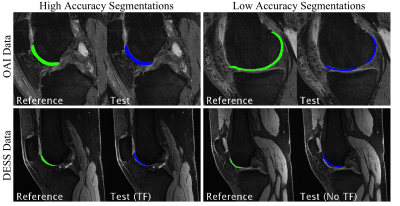
The ‘baseline knowledge’ from the OAI segmentation was used to create a transfer learning (TF) model for a more-recent implementation of 3D DESS sequence acquired on GE Healthcare scanners. Unlike the 11-minute OAI sequence, this 5-minute DESS sequence acquires echoes separately and uses complementary information to highlight or suppress fluid and to create 3D T2 maps. It utilized a 16-channel flexible knee coil (relevant parameters: Matrix=384x320, 80 slices, 2x2 parallel imaging, slice-thickness=1.5mm, FOV=16cm). 6 patients were used for training, 2 for validation, and 2 for testing (examples given in Figure 2). For comparison, the limited-training DESS dataset was also tested on a 3D U-Net trained without transfer learning (No TF model).
Segmentation accuracies for OAI DESS and limited-training DESS datasets were evaluated using Dice coefficients, volumetric overlap error (VOE), and volume difference (VD) measurements.
Results
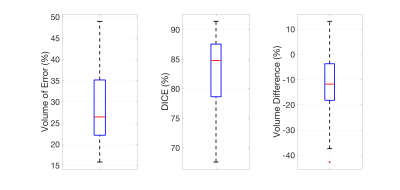
The OAI DESS segmentation accuracy (mean ± standard deviation) with a 3D U-Net model was: VOE = 28.9% ± 10.2%, Dice = 82.7% ± 7.3%, and VD = -13.2% ± 14.2% (seen with box-plots in Figure 3).
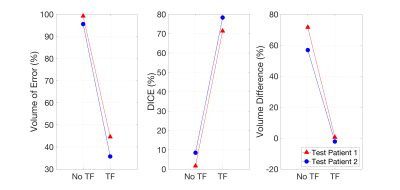
For the limited-training DESS datasets, the TF model demonstrated a dramatic improvement over the model without transfer learning (No TF) in both test cases (test 1, test 2): VOE = (99.2%, 95.6%) improved to (44.6%, 35.7%), Dice = (1.6%, 8.4%) improved to (71.3%, 78.3%), VD = (71.6%, 56.9%) improved to (0.6%, -2.2%) (seen in Figure 4).
Discussion
The 3D U-Net segmentation model for the baseline OAI data outperformed automated femoral cartilage segmentation methods published previously7. The advantages of this network included 3D convolutions and the presence of dropout and batch normalization layers, which made the model more generalizable. Dropout layers randomly disable network weights to force the non-disabled weights to better adapt to the training, while batch normalization ensures that layer weights are non-zero.
While automated-segmentation methods using large publically-available databases such as the OAI and SKI10 have been proposed previously, this study demonstrated that transfer learning can automate segmentation in routinely used 3D research sequences using only 6 training datasets7,10. Such transfer learning methods are important because while thousands of cases are available from the 10-year-old OAI DESS protocol, current DESS implementations offer 3D T2 maps and reduced scan times.
The TF segmentation consistently underestimated cartilage volume (Figure 5), possibly because the OAI training data consist of patients with moderate to advanced OA, which is characterized by cartilage loss. To improve the segmentation model, data augmentation methods could be utilized to simulate unique data from existing data to train the algorithm.
Conclusion
We have demonstrated an automated CNN-based knee cartilage segmentation method that utilizes the wealth of OAI knee data for initial training followed by transfer learning using limited data from faster, newer and more practical 3D sequences that are being used in current studies.Acknowledgements
Research support provided by NIH AR0063643, NIH EB002524, NIH AR062068, NIH EB017739, NIH EB015891, and GE Healthcare.References
1. Heidari B. Knee osteoarthritis prevalence, risk factors, pathogenesis and features: Part I. Caspian J Intern Med. 2011;2(2):205-212.
2. Quatman CE, Hettrich CM, Schmitt LC, Spindler KP. The clinical utility and diagnostic performance of magnetic resonance imaging for identification of early and advanced knee osteoarthritis: a systematic review. Am J Sports Med. 2011;39(7):1557-1568. doi:10.1177/0363546511407612.
3. Bloecker K, Wirth W, Guermazi A, Hitzl W, Hunter DJ, Eckstein F. Longitudinal change in quantitative meniscus measurements in knee osteoarthritis—data from the Osteoarthritis Initiative. Eur Radiol. 2015;25(10):2960-2968. doi:10.1007/s00330-015-3710-7.
4. Eckstein F, Burstein D, Link TM. Quantitative MRI of cartilage and bone: degenerative changes in osteoarthritis. NMR Biomed. 2006;19(7):822-854. doi:10.1002/nbm.1063.
5. Avendi MR, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Medical Image Analysis. 2016;30(Supplement C):108-119. doi:10.1016/j.media.2016.01.005.
6. Brosch T, Tang LYW, Yoo Y, Li DKB, Traboulsee A, Tam R. Deep 3D Convolutional Encoder Networks With Shortcuts for Multiscale Feature Integration Applied to Multiple Sclerosis Lesion Segmentation. IEEE Transactions on Medical Imaging. 2016;35(5):1229-1239. doi:10.1109/TMI.2016.2528821.
7. Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magnetic Resonance in Medicine. 2017. doi:10.1002/mrm.26841.
8. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Miccai. 2015:234–241. doi:10.1007/978-3-319-24574-4_28.
9. Peterfy CG, Schneider E, Nevitt M. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis Cartilage. 2008;16(12):1433–1441. doi:10.1016/j.joca.2008.06.016.
10. Shan L, Zach C, Charles C, Niethammer M. Automatic atlas-based three-label cartilage segmentation from MR knee images. Medical Image Analysis. 2014;18(7):1233–1246. doi:10.1016/j.media.2014.05.008.
Figures