0189
Accurate and Efficient QSM Reconstruction using Deep Learning1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Harvard Medical School, Boston, MA, United States, 3Radiology, Stanford University, Stanford, CA, United States
Synopsis
Quantitative Susceptibility Mapping (QSM) is a powerful MRI technique to quantify susceptibility changes and reveal pathology such as multiple sclerosis (MS) lesions and demyelination. QSM reconstruction is very challenging because it requires solving an ill-posed deconvolution and removing the effects of a dipole kernel on tissue phases to obtain susceptibility. To address the limitations of existing QSM reconstruction methods in accuracy, stability and efficiency, an iteration-free data-driven QSM reconstruction is proposed that trains a deep learning model to approximate COSMOS QSM quantification from acquired signals and pre-processed phases. Cross-validated on in-vivo datasets with 15 single direction QSM scans and 3 COSMOS QSM results from 3 healthy subjects, the proposed deep learning method achieves accurate QSM reconstruction, outperforming state-of-the-art methods across various metrics. The deep learning solution is also faster than iterative reconstruction by several orders of magnitude, which enables broader clinical applications.
Purpose
Quantitative Susceptibility Mapping (QSM) is a powerful MRI technique that quantifies the susceptibility distribution from the measured phase changes. It quantifies chemical identification and reveals pathologies such as iron in multiple sclerosis (MS) lesions, calcification, demyelination, and other pathophysiological susceptibility changes. QSM reconstruction is a challenging ill-posed inverse problem to de-convolve the dipole kernel’s effects on the tissue phase and obtain the susceptibility maps.
Existing QSM reconstruction is limited because:
* 1) The ill-posed deconvolution is under-determined and can only be approximated by enforcing pre-defined regularizations;
* 2) Inaccurate deconvolution may result in large errors that appear as streaking artifacts in the reconstructed QSM images;
* 3) Iterative optimization is used with regularization which is time consuming and difficult to tune.
In this work, we propose a deep-learning-based QSM reconstruction algorithm to overcome these limitations.
Method
- 1. Deep Learning Model Details
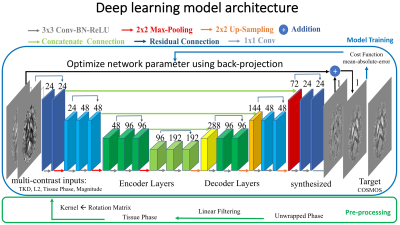
Here we propose an iteration-free reconstruction method using deep learning that enables more accurate and efficient QSM reconstruction. A deep network is trained to generate the QSM reconstruction from the acquired signals and initial estimations of susceptibility map using simpler closed-form estimation methods: 1) magnitude images, 2) raw tissue phase using Laplacian Boundary Value (LBV) background removal from MEDI toolbox, 3) direct estimation from Truncated k-space Division (TKD), 4) closed-form L2 estimation. These 4 different types of images are taken as inputs to the neural network.
The proposed network structure is shown in Figure 1. Symmetric concatenated connections are used to preserve high-resolution QSM information during the encoding-decoding process. L1 cost function and ADAM optimizer are used in the deep learning training.
- 2. Datasets
We validated the method using in-vivo QSM datasets. Fifteen 3D-GRE scans were acquired from three healthy subjects at 3T (5 orientations for each subject, matrix size 224x224, 1mm isotropic resolution, TE=25, single echo at bandwidth=100Hz/pixel, TR=33, FA=15). For ground-truth QSM quantification, we used the COSMOS method as the target reference output, which quantifies more accurate QSM through oversampling with multiple orientations.
- 3. Evaluation
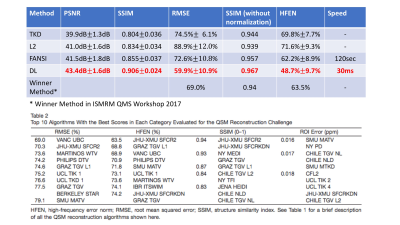
To evaluate the model, Leave-one-out cross-validation (LOOCV) was used in which the result for each subject is generated using the model trained on 10 scans from the other 2 subjects to avoid overfitting and provide a better evaluation of model performance on new datasets. We compared the proposed method with other QSM reconstruction methods: 1) Truncated k-space division (TKD), 2) L2 regularized closed-form deconvolution, 3) Nonlinear MEDI (state-of-the art QSM algorithm, accelerated using FANSI). Several similarity metrics, including PSNR, RMSE, SSIM and HFEN, were used to compare the reconstruction results of different methods to COSMOS ground-truth. The selection of the metrics followed the ones used in the 2016 ISMRM QSM workshop.
- 4. Computation platform
The deep learning method was trained and evaluated using the Keras+Tensorflow framework on a Linux server with 1 NVIDIA 1080TI GPU.
Results
From quantitative metrics shown in Table 1, the proposed deep-learning-based QSM reconstruction shows significant improvements over existing methods. In addition, the proposed method is faster by several orders of magnitude than MEDI/FANSI, the state-of-the-art QSM methods that uses pre-defined regularization and requires iterative optimization.
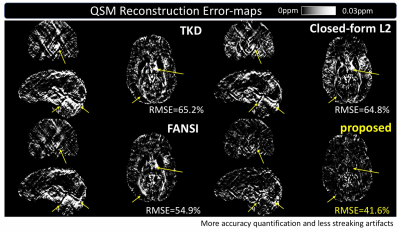
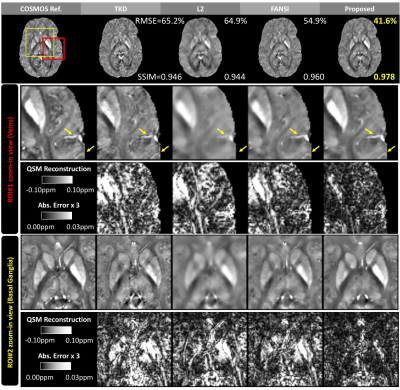
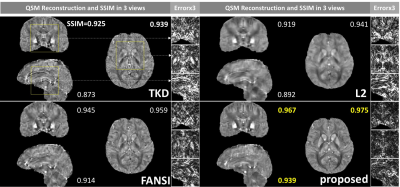
The visual comparison in Figure 2 visualize the error-maps of different methods on one subject in 3 (axial/coronal/sagittal) views and demonstrates the significant reduction of streaking artifacts.
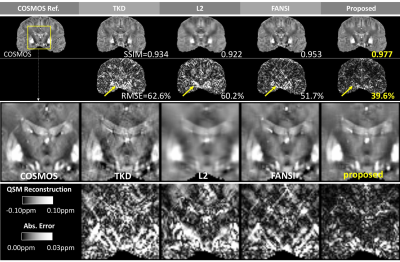
Figure 3 shows the results on another subject and the zoom-in views visualize how the proposed method reduced artifacts and improved quantification of susceptibility at Basal Ganglia.
Figure 4 and Figure 5 demonstrates the reduced error and finer details around both Basal Ganglia and veins. Detailed accuracy metrics (SSIM and/or RMSE) are also shown in figure.
Discussion
Compared with existing methods, the proposed method demonstrates significant improvements in accuracy and efficiency. As shown in Figure 2-5, the reconstruction improvement is especially well noticed around basal ganglia and veins, showing more accurate quantification and better suppressed streaking artifacts.
Another advantage is that the end-to-end deep learning pipeline can be flexibly improved and fine-tuned for certain performance metrics or different purposes. For example, HFEN can be used in part of the cost function when high-frequency performance is more important for specific QSM application.
The future direction for this study is to collect more in vivo datasets on both healthy controls and patients for more extensive evaluation.
Conclusion
Here we proposed an iteration-free method for QSM reconstruction using deep learning. Compared with the state-of-the-art QSM reconstruction methods, the proposed deep learning approach shows superiority in both accuracy and efficiency.
Acknowledgements
No acknowledgement found.References
1) Wang, Yi, and Tian Liu. "Quantitative susceptibility mapping (QSM): decoding MRI data for a tissue magnetic biomarker." Magnetic resonance in medicine 73.1 (2015): 82-101.
2) Wharton, Sam, Andreas Schäfer, and Richard Bowtell. "Susceptibility mapping in the human brain using threshold‐based k‐space division." Magnetic resonance in medicine 63.5 (2010): 1292-1304.
3) Bilgic, Berkin, et al. "Fast image reconstruction with L2‐regularization." Journal of Magnetic Resonance Imaging 40.1 (2014): 181-191.
4) de Rochefort, Ludovic, et al. "Quantitative susceptibility map reconstruction from MR phase data using bayesian regularization: validation and application to brain imaging." Magnetic Resonance in Medicine 63.1 (2010): 194-206.
5) C Milovic et al ISMRM’17
Figures