0165
Enhancing In Vivo Hyperpolarized 13C Chemical Shift Imaging by an Iterative Reconstruction1Chemical & Biological Physics, Weizmann Institue, Rehovot, Israel
Synopsis
Hyperpolarized 13C magnetic resonance spectroscopic imaging (MRSI) is a powerful metabolic technique, but it’s challenged by a rapid and irreversible decay of the signal that usually compromises its achievable spatial resolution. In this work we explore a way to improve this by utilizing a priori anatomical information derived from 1H MRI. Enhanced HP-MRSI implementations based on Spectroscopy with Linear Algebraic Modeling (SLAM) were thus assayed, to enhance HP-MRSI’s spatial resolution without compromising SNR. 13C experiments were performed in-vivo and pyruvate/lactate images reconstructed for physiological compartments by SLAM; we compare these results to those arising by traditional Fourier analyses.
Introduction
DNP [1] increases the 13C spin polarization to the point of enabling in vivo MR imaging and spectroscopy studies of labeled metabolites with high signal-to-noise ratio (SNR) [1,2]. This has allowed tracking metabolism in-vivo, usually by injecting labeled 13C1-pyruvate and mapping its conversion into 13C1-lactate MRSI [3,4]. These data need in turn be collected at multiple time points to probe metabolic rates. The need to probe three spatial and one spectral dimension multiple times over the limited lifetimes of a hyperpolarized substrate, limits the resolution with which any of these variables can be examined. This work explores the possibility of improving spatial aspects of these acquisitions, by applying a model-based reconstruction relying on a priori segmentations of anatomical scans, arising from 1H MRI. This will in turn assume that the metabolic signals are spatially uniform with certain physiological compartments; although this may not be a valid assumption if attempting to detect an unkowon feature, we found it appealing for rodent experiments where organs/tumors are small, and compromises made by 13C MRSI lead to spillover effects. The approach we developed for exploring this is based on Spectroscopy with Linear Algebraic Modeling (SLAM) [5], which derived from similar limitations found in thermal 1H MRSI. Ways in which SLAM can be exploited include enhancing the 13C MRSI’s spatial resolution, improving SNR while maintaining spatial resolution, or reducing the number of k-samples and thereby preserving polarization for better kinetic characterizations. In this study we focus on the first strategy, to bring a sharper compartmentalization and better match between the 13C and the 1H MRI anatomies to DNP MRSI.Methods
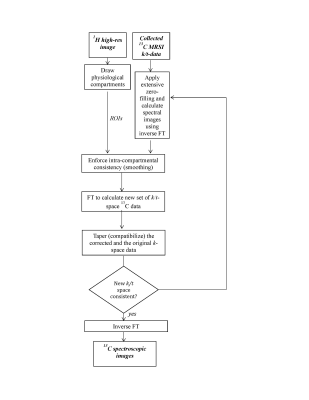
A SLAM-based reconstruction algorithm was developed, enforcing consistency between a 1H Region-of-Interest (ROI) and uniformity in the 13C spectral intensities within that ROI. Inspired by iterative POCS [6,7], our algorithm operated as follows (Figure 1): (1) Interpolated 13C spectral images were reconstructed using the k-space data and zero-filling; (2) Uniformity within the segmented high-resolution 1H ROI was enforced on the 13C images by replacing the 13C pixels in the ROI with either a simple or a weighted average of all its members; (3) FT was applied on these images to calculate a “corrected” set of k-space data; (4) This new k-space data was “tapered” to ensure a smooth transition between the original and the corrected k-space data. (5) Steps 1-4 were repeated until the iteratively calculated k-space data showed consistency. This algorithm was tested in vitro on a 7T Agilent scanner using 1H volume and 13C surface coils, and in vivo by focusing on the placenta of Wistar pregnant rats. Six such cases were studied at days 17-21 of gestation, on a 4.7T Bruker Biospec scanner. 13C1-pyruvic acid was in all cases polarized with 15 mM Ox63 in Hypersense® operating at 94 GHz and 1.4 K. All MRSI experiments relied on home-written centric k-sampled chemical shift imaging sequences [8].Results
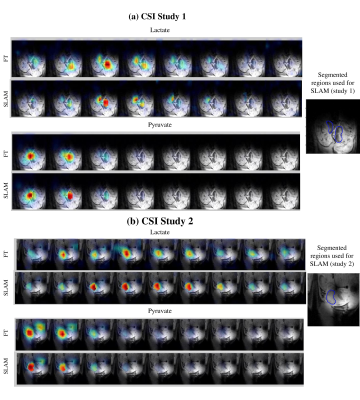
In vitro SLAM results shown in Figure 2 on a two-compartment 1H phantom containing water/DMSO, and on a tube where hyperpolarized 13C1-pyruvic acid was injected. In both cases SLAM shows improvements over conventional FT in terms of spatial definition and SNR. While these studies for which SNR was high and the ROI’s were known could implement step #2 above by taking a simple average of the voxels’ intensities, this failed for in vivo cases. For these, step #2 had to incorporate a weighted Gaussian average based on distance between the 13C pixels, to ensure a stable convergence of the SLAM algorithm. Figure 3 shows representative outcomes of this procedure, for in vivo studies exploring the placental conversion of Pyruvate into Lactate. These results show that the SLAM reconstruction can improve SNR and spatial resolution, even in these challenging abdominal conditions. An important requirement of these experiments is also the maintenance of kinetic faithfulness. The comparisons between SLAM and FT-based metabolite kinetic curves presented in Figure 4, confirm that this feature is also preserved.Conclusions
A reconstruction algorithm based on a-priori segmentations of physiological compartments, has been developed and tested in vitro and in-vivo on hyperpolarized data. Results on phantoms show definition and SNR advantages of SLAM over conventional FT. Also the in vivo results proved superior in these aspects, while maintaining consistency with the FT-derived kinetics. Extensions to a larger number of in vivo studies is in progress to assess this approach’s generality.Acknowledgements
Funding from the Israel Science Foundation (#2508/17), EU ERC-2016-PoC grant # 751106, Minerva Foundation - Germany (#712277), Kimmel Institute for Magnetic Resonance & Perlman Family Foundation (Weizmann), are gratefully acknowledged. We are grateful to Drs. Y. Zhang and P. Bottomley (Johns Hopkins) for discussions.References
[1] J.H. Ardenkjaer-Larsen, B. Fridlund, A. Gram, G. Hansson, L. Hansson, M.H. Lerche, R. Servin, M. Thaning, K. Golman, Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR, Proc. Natl. Acad. Sci. (2003).
[2] J. Kurhanewicz, D.B. Vigneron, K. Brindle, E.Y. Chekmenev, A. Comment, C.H. Cunningham, R.J. DeBerardinis, G.G. Green, M.O. Leach, S.S. Rajan, R.R. Rizi, B.D. Ross, W.S. Warren, C.R. Malloy, Analysis of Cancer Metabolism by Imaging Hyperpolarized Nuclei: Prospects for Translation to Clinical Research, Neoplasia. (2011).
[3] K. Golman, R. in ’t Zandt, M. Thaning, Real-time metabolic imaging., Proc. Natl. Acad. Sci. U. S. A. 103 (2006) 11270–5.
[4] K.M. Brindle, Imaging Metabolism with Hyperpolarized 13C-Labeled Cell Substrates, J. Am. Chem. Soc. 137 (2015) 6418–6427.
[5] Y. Zhang, R.E. Gabr, M. Schär, R.G. Weiss, P.A. Bottomley, Magnetic resonance Spectroscopy with Linear Algebraic Modeling (SLAM) for higher speed and sensitivity, J. Magn. Reson. (2012).
[6] Y. Xu, E. Mark Haacke, Partial fourier imaging in multi-dimensions: A means to save a full factor of two in time, J. Magn. Reson. Imaging. (2001).
[7] E.M. Haacke, E.D. Lindskogj, W. Lin, A fast, iterative, partial-fourier technique capable of local phase recovery, J. Magn. Reson. (1991).
[8] E. Adalsteinsson, P. Irarrazabal, S. Topp, C. Meyer, A. Macovski, D.M. Spielman, Volumetric spectroscopic imaging with spiral-based k-space trajectories, Magn. Reson. Med. (1998).
Figures