0157
Automatic Removal of Ghosting Artifacts from MR Spectra using Deep Learning1Depts. Radiology and Biomedical Research, University of Bern, Bern, Switzerland
Synopsis
Ghosting artifacts in clinical MR spectroscopy are problematic since they superimpose with metabolites and lead to inaccurate quantification. Here, we make use of “Deep Learning” (DL) methods to remove ghosting artifacts in MR spectra of human brain. The DL method was trained on a huge database of simulated spectra with and without ghosting artifacts, which represent complex variants of ghost-ridden spectra, transformed to time-frequency spectrograms. The trained model was tested on simulated and in-vivo spectra. The preliminary results for ghost removal show potential in simulated and in-vivo spectra, but need further refinement and quantitative testing.
Introduction
Ghosting artifacts in spectroscopy are problematic since they superimpose with metabolites and lead to inaccurate quantification. It has previously been shown1 that detection of ghosting artifacts based on deep learning (DL) is possible. In this study, the previous approach is extended to remove ghosting artifacts to arrive at undistorted spectra using stacked-autoencoders, a type of DL method that has previously been used to remove noise from images2–4.Methods
MR spectra: Brain spectra were simulated as in Ref [1] for two cohorts of spectra: Group‑1 consisted of spectra with and without ghosts with identical metabolite content, varied in linewidth (LW) and SNR. Group‑2 included variations in metabolite concentration, baseline and lipid contributions. 1D spectra were converted to a 2D time-frequency representation using piecewise Fourier-transformation (window size of 56 points (~14ms), overlap interval of 48 points and Hanning windowing) to get complex spectrograms with 56 frequency bins and 250 time stamps. In vivo spectra with ghosting artifacts had been obtained with a diffusion-weighted STEAM sequence (TE 37 ms) at 3T in gray matter from 13 healthy subjects.
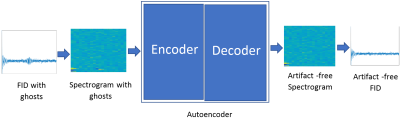
Ghost removal: The standard stacked-autoencoder architecture is composed of encoding and decoding. The encoder layer consists of 2D-convolutional filters and activation functions followed by a max-pooling filter. The decoder network has a reverse process and consists of up-sampling layers. Here, Stacked What-Where Auto Encoders (SWWAEs)5 built on residual blocks6 were implemented and tested for removing ghosting artifacts from spectra. Spectrograms with ghosting artifacts (27500) randomly selected from both Group-1 and Group-2 were used as input and corresponding spectrograms without artifacts were given as ground truth data (Figure-1). The trained SWWAE was tested on 5000 independent simulated spectrograms and 21 in vivo spectrograms with ghosting artifacts to predict the artifact free spectrogram. For plotting and quantitative judgment of the similarity between ground truth and the reconstructed spectra the spectra were normalized using the L-2 norm. The root-mean-squared error (RMSE) between the normalized predicted and the normalized restored spectrum was calculated for all test cases. The mean RMSE over all spectra and separately for the best and worst SNR cases used (SNR-10 and SNR-100) were given as output measures.
Results
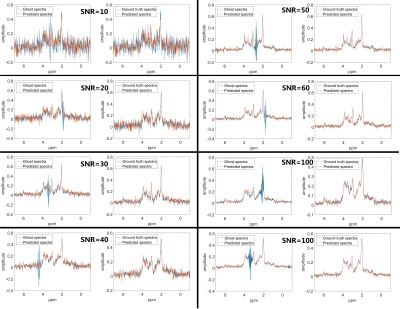
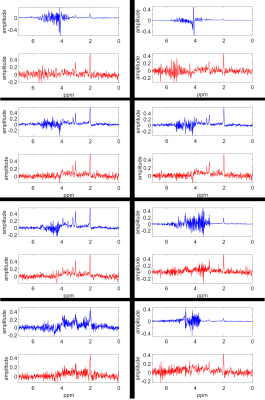
With the proposed SWWAE, it was possible to restore ghost-distorted spectra on simulated cases. Figure-2 shows sample spectra with ghosting artifacts in comparison to the restored spectra and the ground truth for different SNRs and LWs. The average RMSE was 0.026 if determined for all test spectra; ranging between 0.052 for those with SNR-10, to 0.017 for those with SNR-100, respectively. As can be judged from Figure-2, these mean deviations are smaller than the noise level. Figure-3 illustrates the restoration of spectra for in-vivo cases, where clearly very large ghosting artifacts have been eliminated, but where it is also evident that some distortions remain and the restored peaks will have to be scrutinized for other distortions.Discussion and Conclusion:
It is possible to use DL methods not only to detect ghosting artifacts in in-vivo MRS but also to remove them. Initial results show promising performance. Even though the simulations covered as much of the expected variance as possible, the DL model did not perform perfectly in removing the artifacts, in particular for in-vivo cases. However, judgment of success for ghost removal has so far only been based on RMSE for the simulated cases and visual appearance for in vivo spectra. This will have to be extended to quantitative evaluations, particularly since the DL restoration step may easily distort parts of the spectrum that are not affected by the overlaid artifact, and apparent success in ghost removal may come at a cost in terms of accuracy for the evaluation of small metabolite contributions throughout the spectrum. In addition, the in-vivo spectra were all of the same type and from healthy brain tissue. Hence this initial trial has to be extended to other acquisition parameters and pathological spectral alterations. However, the use of DL techniques in spectroscopy is a wide and open field and the current contribution can only scratch the surface.Acknowledgements
This research was carried out in the framework of the European Marie-Curie Initial Training Network, ‘TRANSACT’, PITN-GA-2012-316679, 2013-2017 and also supported by the Swiss National Science FoundationReferences
1. Kyathanahally, S. P., Doering, A. & Kreis, R. Ghostbusters for MRS: Automatic Detection of Ghosting Artifacts using Deep Learning. in Proceedings of the 25th Annual Meeting ISMRM 5479 (2017).
2. Xie, J., Xu, L. & Chen, E. Image denoising and inpainting with deep neural networks. in Proceedings of the 25th International Conference on Neural Information Processing Systems 341–349 (2012).
3. Gondara, L. Medical image denoising using convolutional denoising autoencoders. in 2016 IEEE 16th International Conference on Data Mining Workshops (ICDMW) 241–246 (2016). doi:10.1109/ICDMW.2016.0041
4. Mao, X.-J., Shen, C. & Yang, Y.-B. Image restoration using convolutional auto-encoders with symmetric skip connections. CoRR abs/1606.0, 1–17 (2016).
5. Zhao, J., Mathieu, M., Goroshin, R. & LeCun, Y. Stacked What-Where Auto-encoders. arXiv:1506.02351 1, 1–12 (2015).
6. Chollet, F. Keras. GitHub, https://github.com/fchollet (2016).
7. Clevert, D.-A., Unterthiner, T. & Hochreiter, S. Fast and accurate deep network learning by Exponential Linear Units (ELUs). CoRR abs/1511.0, (2015).
8. Zeiler, M. D. ADADELTA: An adaptive learning rate method. CoRR abs/1212.5, (2012).
Figures