0150
Ultrahigh spatiotemporal-resolution fMRI reveals distinct brain-wide functional networks of different hippocampal subfields1BRIC, UNC at Chapel Hill, Chapel Hill, NC, United States, 2Department of Psychology, UNC at Chapel Hill, Chapel Hill, NC, United States
Synopsis
The hippocampal formation consists of distinct subfields, which contribute to different aspects of memory function, and exhibit different brain network topologies. However, the brain-wide resting-state functional connectivity of hippocampal subfields in human remains poorly understood mainly due to technical limitations. Previous efforts to identify hippocampal subfield functional networks compromised either spatial resolution, spatial coverage or temporal resolution. We have developed a new approach, named Partition-encoded Simultaneous Multi-slab (PRISM), capable of acquiring ultrahigh isotropic resolution images while maintaining the acceleration capability. Our results of resting-state functional connectivity at 7T reveal distinct brain-wide functional networks associated with different hippocampal subfields.
Introduction
Hippocampus plays a major role in learning and memory, and it is the earliest affected structure in Alzheimer's disease (AD). Although the subfield volumetry has been suggested to yield more accurate diagnosis than total hippocampal volumetry1-3, the subfield volumetry is not specific to AD4-7 and its sensitivity is still inconclusive8. Another method is network-based approaches which take into account the brain-wide functional connectivity (FC) rather than local structural change. However, studying the brain-wide resting-state functional connectivity (rsFC) of hippocampal subfield is technically challenging. Ultrahigh-resolution functional imaging is required due to the small size of hippocampal subfields. Meanwhile, images encompassing the entire brain and acquisition within a few seconds are needed. Previous attempts compromised either spatial resolution (>2 mm)9, spatial coverage (<1/2 brain volume)10 or temporal resolution (>3 s)11. We recently developed a novel method named Partition-encoded Simultaneous Multi-slab (PRISM) imaging to mitigate the aforementioned challenges, achieving whole-brain fMRI with 1-mm isotropic resolution in 2 seconds12. In this study, functional architecture of cortical-hippocampal subfield networks are revealed using PRISM.Method
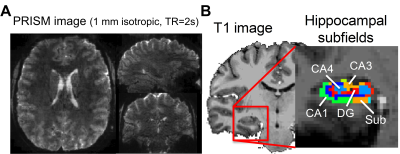
We recruited 8 healthy adults aged 22 to 41 years old (31.8±6.6 years; 4 males and 4 females). MR images were acquired using a Siemens 7T scanner. High-resolution MPRAGE images were acquired initially, followed by two short PRISM scans with forward and reversed phase-encoding directions. We then collected an 8-minute resting-state scans using the PRISM sequence. The spatial resolution was 1 mm isotropic and field-of-view (FOV) of 112 x 152 x 175 mm3. The total number of coronal slabs was 35, TE=22 ms and TR=2s, SMS factor=7, in-plane acceleration factor=2 and the number of through-slab PAE=5. The concatenation of slice GRAPPA13 and in-plane GRAPPA14 was used for image reconstruction. The ‘topup’ command in FSL was used to correct EPI distortion. The time series were motion corrected using FSL15. The motion-corrected data were decomposed into a number of independent components by MELODIC16. The noise components were identified and removed automatically by FIX17. The time series were low-pass filtered at 0.1 Hz. The EPI images were coregistered onto individual anatomical image by affine transformation before being coregisterd onto the MNI template by ANTs18.To perform seed-based connectivity analysis, we used Freesurfer 6.0 for automatic hippocampal subfield segmentation for individual and template brains19. Four hippocampal subfields including subiculum, cornu ammonis 1(CA1), CA3 and CA4 and dentate gyrus (DG) were used as seed regions, separately. The seed-based resting-state FC (rsFC) at the individual level was calculated using Pearson correlation before being Fisher Z-transformed. The results of Fisher Z were coregistered onto the template brain. Correction for multiple comparison was performed using the FSL command ‘easythresh’ by setting the voxelwise p < 0.05 and cluster-wise p < 0.05.Results
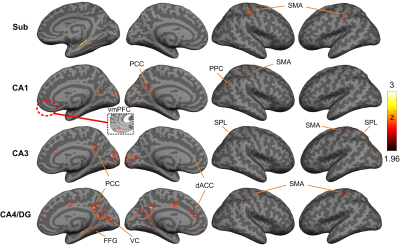
Figure 1A and 1B showed the reconstructed PRISM images and the hippocampal subfields, respectively. The group averaged resting-state functional networks were morphed onto the cortical surface of the template for better visualization (see Figure 2). All subfields showed the connections with sensorimotor area (SMA). CA1 also showed connectivity with ventral medial prefrontal cortex (vmPFC) and posterior parietal cortex (PPC). CA1, CA3 and CA4/DG all demonstrated significant connectivity with posterior cingulated cortex (PCC). Additionally, CA3 and CA4/DG showed connectivity with anterior cingulated cortex (ACC). The cingulate cortex is known to be involved in learning and memory process20-21. Moreover, CA3 showed connections with bilateral superior parietal lobule, which is critical for working memory. We also observed that CA4/DG showed stronger connectivity with primary visual cortex (VC) and fusiform gyrus (FFG) relative to other hippocampal subfields.Discussion
This study is, to the best of our knowledge, the first to demonstrate brain-wide functional networks of different hippocampal subfields. More specifically, most of the regions involved in the subfield networks were supported by the early findings, such as cingulated cortex and parietal cortex. Prior study of subfield volumetry have reported that CA1 is associated with memory consolidation while CA3 and DG are associated with learning22. Our result of CA1 network was consistent with prior finding, exhibiting significant connectivity with vmPFC and PPC which are implicated in memory consolidation23-24. Likewise, the PCC involved in the CA4/DG network has been linked with learning25. Our results further suggested that primary visual area and fusiform gyrus are involved in the CA4/DG network. This finding is consistent with prior studies of pattern separation that require processing of visual details26. In conclusion, the unprecedented feature of PRISM imaging will empower the field to investigate the cortical-hippocampal subfield networks mediating healthy versus pathological aging, thereby serving as a potential early biomarker of AD.Acknowledgements
No acknowledgement found.References
1. Lo, R.Y., et al., Longitudinal change of biomarkers in cognitive decline. Arch Neurol, 2011. 68(10): p. 1257-66.
2. La Joie, R., et al., Hippocampal subfield volumetry in mild cognitive impairment, Alzheimer's disease and semantic dementia. Neuroimage Clin, 2013. 3: p. 155-62.
3. Jack, C.R., Jr., et al., Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol, 2013. 12(2): p. 207-16.
4. Sabattoli, F., et al., Hippocampal shape differences in dementia with Lewy bodies. Neuroimage, 2008. 41(3): p. 699-705.
5. Lindberg, O., et al., Hippocampal shape analysis in Alzheimer's disease and frontotemporal lobar degeneration subtypes. J Alzheimers Dis, 2012. 30(2): p. 355-65.
6. Kril, J.J., et al., Patients with vascular dementia due to microvascular pathology have significant hippocampal neuronal loss. J Neurol Neurosurg Psychiatry, 2002. 72(6): p. 747-51.
7. Gemmell, E., et al., Hippocampal neuronal atrophy and cognitive function in delayed poststroke and aging-related dementias. Stroke, 2012. 43(3): p. 808-14.
8. Maruszak, A. and S. Thuret, Why looking at the whole hippocampus is not enough-a critical role for anteroposterior axis, subfield and activation analyses to enhance predictive value of hippocampal changes for Alzheimer's disease diagnosis. Front Cell Neurosci, 2014. 8: p. 95.
9. de Flores, R., et al., Intrinsic connectivity of hippocampal subfields in normal elderly and mild cognitive impairment patients. Hum Brain Mapp, 2017.
10. Yassa, M.A., et al., Age-related memory deficits linked to circuit-specific disruptions in the hippocampus. Proc Natl Acad Sci U S A, 2011. 108(21): p. 8873-8.
11. Bergmann, E., et al., The Organization of Mouse and Human Cortico-Hippocampal Networks Estimated by Intrinsic Functional Connectivity. Cereb Cortex, 2016. 26(12): p. 4497-4512.
12. Chang, W.T. and W. Lin. Fast imaging with ultrahigh isotropic resolution using partition-encoded simultaneous multi-slab (PRISM). in International Society for Magnetic Resonance in Medicine. 2018. #8446. Paris, France.
13. Setsompop, K., et al., Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn Reson Med, 2012. 67(5): p. 1210-24.
14. Griswold, M.A., et al., Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med, 2002. 47(6): p. 1202-10.
15. Smith, S.M., et al., Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage, 2004. 23 Suppl 1: p. S208-19.
16. Beckmann, C.F., et al., Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci, 2005. 360(1457): p. 1001-13.
17. Salimi-Khorshidi, G., et al., Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage, 2014. 90: p. 449-68.
18. Avants, B.B., et al., The Insight ToolKit image registration framework. Front Neuroinform, 2014. 8: p. 44.
19. Iglesias, J.E., et al., A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage, 2015. 115: p. 117-37.
20. Cingulate binds learning. Trends Cogn Sci, 1997. 1(1): p. 2.
21. Frankland, P.W., et al., The involvement of the anterior cingulate cortex in remote contextual fear memory. Science, 2004. 304(5672): p. 881-3.
22. Mueller, S.G., et al., Evidence for functional specialization of hippocampal subfields detected by MR subfield volumetry on high resolution images at 4 T. Neuroimage, 2011. 56(3): p. 851-7.
23. Nieuwenhuis, I.L. and A. Takashima, The role of the ventromedial prefrontal cortex in memory consolidation. Behav Brain Res, 2011. 218(2): p. 325-34.
24. Liu, Y., et al., Memory consolidation reconfigures neural pathways involved in the suppression of emotional memories. Nat Commun, 2016. 7: p. 13375.
25. Sutherland, R.J., I.Q. Whishaw, and B. Kolb, Contributions of cingulate cortex to two forms of spatial learning and memory. J Neurosci, 1988. 8(6): p. 1863-72.
26. Bakker, A., et al., Pattern separation in the human hippocampal CA3 and dentate gyrus. Science, 2008. 319(5870): p. 1640-2.
Figures