0133
Histological Validation of in-vivo VERDICT MRI for Prostate using 3D Personalised Moulds1Centre for Medical Image Computing, University College London, London, United Kingdom, 2Centre for Medical Imaging, University College London, London, United Kingdom, 3Research Department for Tissue & Energy, University College London, London, United Kingdom, 4Department of Pathology, University College London Hospital, London, United Kingdom, 5Division of Surgery and Interventional Science, University College London, London, United Kingdom
Synopsis
VERDICT analysis can successfully distinguish benign from malignant prostate tissue in-vivo showing promising results as a cancer diagnostic tool. However, the accuracy with which model parameters reflect the underlying tissue characteristics is unknown. In this study, we quantitatively compare the intracellular, extracellular-extravascular and vascular volume fractions derived from in-vivo VERDICT MRI against histological measurements from prostatectomies. We use 3D-printed personalised moulds designed from in-vivo MRI that help preserve the orientation and location of the gland and aid histological alignment. Results from the first samples using the 3D mould pipeline show good agreement between in-vivo VERDICT estimates and histology.
INTRODUCTION
VERDICT aims to provide microstructural information from solid cancer tumours non-invasively using mathematical models linking the diffusion MRI signal to microscopic tissue features1. The VERDICT model for prostate cancer characterisation shows promising results in differentiating benign and cancerous tissue and correlates with different Gleason patterns1,2. However, few attempts have been made to validate the clinical VERDICT for prostate using histological ground truth3 and rigorous validation of the in-vivo parameters has never been done.
MRI validation is challenging in matching its registration with histology. During in-vivo imaging, the prostate is deformed (due to voluntary and involuntary body movements) which makes it difficult to match even the corresponding tissue planes. Using personalised-moulds from in-vivo MRI4 with landmarks has demonstrated consistent tissue positioning and minimised misalignment between ex-vivo VERDICT MRI and histological features5. This study goes one step further and uses 3D-personalised-mould, with additional ex-vivo MRI for alignment to aid the registration of in-vivo MRI and histopathology.
The aim of this study is to perform a direct quantitative validation of the in-vivo VERDICT parametric maps for prostate cancer1,6 against histological sections obtained from radical prostatectomies.
METHODS
Imaging
Three different candidates for prostatectomy with a previous clinical multiparametric prostate MRI (mpMRI) were scanned for VERDICT analysis as part of the INNOVATE clinical trial7. For each VERDICT scan, diffusion-weighted (DW) MRI was performed using a 3T scanner (Achieva, Philips Healthcare, Netherlands) as in2,8. The VERDICT model was fitted to the data in each voxel using the AMICO9 framework to obtain the parameter maps as previously described in8. The VERDICT parameters that we are validating here are the volume fractions (vf): intracellular (fIC), extracellular-extravascular (fEES) and vascular (fVASC).
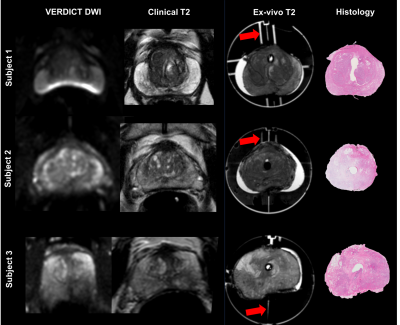
The clinical mpMRI was used to create patient-specific 3D-printed prostate moulds with anatomical landmarks to preserve the original shape of the organ and to be able to identify areas of interest4. After the prostatectomy, prostates were placed in the mould, following the protocol described in4 and scanned again fresh (ex-vivo) in the 3T scanner. We used landmarks (red arrows in Figure 1,3a) in the 3D-mould to select the same slice.
Histological analysis
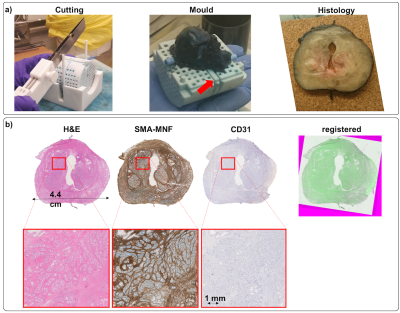
Samples were cut in 5mm sections using the mould guides. From each section, three consecutive 3μm slices were obtained, stained with (1) hematoxylin and eosin (H&E), (2) cytokeratin and smooth muscle actin (MNF-SMA) and (3) CD31 marker (CD31), respectively and digitised (Hamamatsu NanoZoomer) using a 20x objective. Slices were aligned with manual rotation and translation. We segmented SMA-MNF images to obtain the proportion of epithelial cells and CD-31 images for quantifying the vasculature.
We analysed different regions of interest (ROI) in the VERDICT-DWI image and in the corresponding histological slice.
RESULTS
Figure 1 shows the different MRI acquisitions with the corresponding histological slices. Important intra-patient differences between in-vivo scans (clinical vs. VERDICT) highlight the challenge for accurate histological correlation.
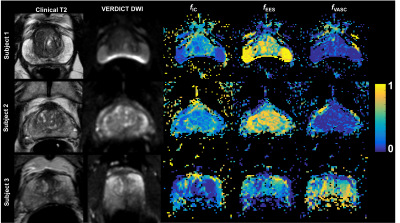
Figure 2 shows the VERDICT maps for the three patients. Maps have been filtered removing voxels with poor fitting using the objective function. As expected, in all samples, prostate peripheral zone has lower fIC than transition zone10.
Figure 3 illustrates for one subject the 3D-mould with the prostate and the corresponding 5mm histological section. Also, it shows the three different stains and the alignment between the histological slices.
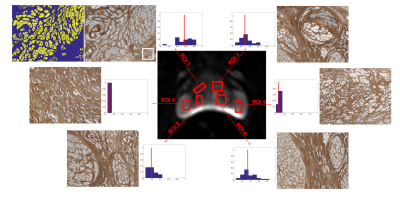
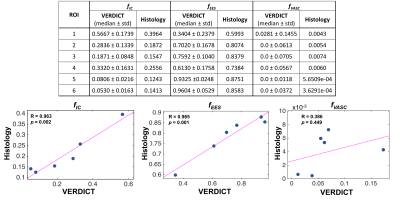
Figure 4 shows the fIC distribution per voxel for different ROIs with corresponding histology. Results reveal that regions with fewer epithelial cells correspond to areas of lower fIC. Table 1 shows the quantitative VERDICT-histology comparison and the correlation for the fIC (r=0.96, p=0.002), fEES (r=0.96, p=0.001) and fVASC (r=0.386, p=0.449). We note that these samples had very low vasculature, hence the close to zero and noisy estimates of fVASC. The rest of vf show agreement with histology.
DISCUSSION
Our study is the first to show that in-vivo VERDICT vf parameters agree well with histology. We used 3D-personalised moulds and ex-vivo scans to aid precise tissue comparison. However, in-vivo images still exhibit prostate deformations and distortions (i.e. bowel gas) that hinder accurate in-vivo to ex-vivo registrations. Future work will improve VERDICT alignment with histology by: (1) utilising the T2 images from the VERDICT acquisition, instead of mpMRI, to create 3D-personalised moulds and (2) using tissue information such as anatomical structures and lesions, prior to registration, to segment regions with similar shrinkage. With better tissue agreement, a more accurate voxel-wise comparison could be performed as in3. More samples are required to further validate all VERDICT parameters.CONCLUSIONS
Early results from the first samples using personalised 3D-printed moulds from MRI show good agreement between in-vivo VERDICT vf estimates and histology.Acknowledgements
This work is funded by a grant PG14-018-TR2 from Prostate Cancer UK. A grant 510419 from the UCLH Biomedical Research Centre supports SP, EPSRC grant EP/N021967/1 supports EP's work on this topic, and EP/M020533/1 and EP/N018702/1 support DCA and DH.References
1. Panagiotaki E, Chan RW, Dikaios N, et al. Microstructural Characterization of Normal and Malignant Human Prostate Tissue With Vascular, Extracellular, and Restricted Diffusion for Cytometry in Tumours Magnetic Resonance Imaging. Investigative Radiology. 2015;50(4):218–227.
2. Johnston E, Bonet-Carne E, Panagiotaki E, et al. Short term repeatability of microstructural (VERDICT) MRI vs. ADC in prostate cancer. International Society for Magnetic Resonance in Medicine (ISMRM). 2016;(24):579.
3. Jacobs JG, Johnston E, Freeman A, et al. Histological validation of VERDICT cellularity map in a prostatectomy case. International Society for Magnetic Resonance in Medicine (ISMRM). 2016; (24):2497.
4. Bourne RM, Bailey C, Johnston EW, et al. Apparatus for Histological Validation of In Vivo and Ex Vivo Magnetic Resonance Imaging of the Human Prostate. Frontiers in Oncology. 2017;7:47.
5. Bailey C, Bourne RM, Siow B, et al. Validation of VERDICT MRI using fresh and fixed prostate specimens with aligned histological slices. International Society for Magnetic Resonance in Medicine (ISMRM). 2017; (25):0868.
6. Panagiotaki E, Walker-Samuel S, Siow B, et al. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res. 2014;74.
7. Johnston E, Pye H, Bonet-Carne E, et al. INNOVATE: A prospective cohort study combining serum and urinary biomarkers with novel diffusion-weighted magnetic resonance imaging for the prediction and characterization of prostate cancer. BMC Cancer. 2016;16(1):816.
8. Bonet-Carne E, Daducci A, Panagiotaki E, et al. Non-invasive quantification of prostate cancer using AMICO framework for VERDICT MR. In: International Society for Magnetic Resonance in Medicine (ISMRM). 2016; (24):3465.
9. Daducci A, Canales-Rodríguez EJ, Zhang H, et al. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. NeuroImage. 2015;105:32–44.
10. McNeal JE. Normal histology of the prostate. The American journal of surgical pathology. 1988;12(8):619–33.
Figures