0132
Do Non-Gaussian diffusion MRI methods improve the detection or specification of cellular alterations following traumatic brain injury?1QMI/NIBIB, National Institutes of Health, Bethesda, MD, United States, 2Henry M. Jackson Foundation, Bethesda, MD, United States, 3NIBIB, National Institutes of Health, Bethesda, MD, United States, 4NICHD, National Institutes of Health, Bethesda, MD, United States, 5APG, Uniformed Services University, Bethesda, MD, United States
Synopsis
Following traumatic brain injury (TBI), numerous microscale cellular alterations appear and evolve with a range of consequences for adverse outcomes and recovery. Diffusion tensor MRI (DTI) has been identified as a potentially sensitive tool for characterizing these changes, but is notably limited in providing specific information about particular cellular alterations and more advanced non-Gaussian frameworks have been developed that may address these limitations. To assess the utility of non-Gaussian modeling for improved detection and specification of TBI-related cellular alterations, we compared DTI, DKI and MAP-MRI in mouse brains following mild TBI and their correspondence to histopathology in the same tissue.
Introduction
Traumatic brain injury (TBI) results in a complex collection of physiological and cellular alterations that are prominent on the microscale, but can escape detection and accurate specification on MRI brain scans especially for mild TBI. Diffusion MRI (dMRI) approaches have been identified as promising for TBI studies given their ability to quantitatively characterize the tissue micro-scale environment1. While Gaussian dMRI approaches (i.e. diffusion tensor imaging, DTI) can be sensitive to such abnormalities2, DTI may not provide enough information to distinguish specific cellular changes. To meet this challenge a number of non-Gaussian dMRI models have been conceived and developed but their potential contribution to TBI research and limitations are not well understood. The objective of the present work is a cross-model evaluation of DTI and non-Gaussian diffusion models to determine the relative advantages and limitations for the detection and specificity of cellular alterations using high-quality ex-vivo DWI data in a mouse model of mild TBI.Methods
Experimental TBI was performed in mice (n=16 TBI, n=4 controls) using standard methods for controlled cortical impact (CCI) with mild settings and brains were obtained at 24-hours, 1week, 4 weeks and 12 weeks after CCI. Following perfusion fixation, brains prepared for imaging and MRI acquisition was performed using a 7T vertical bore MRI scanner with a 10mm linear coil to collect structural T2W and 3D-EPI diffusion weighted image (DWI) volumes with 100 micron isotropic resolution using a multi-shell DWI sampling of 297 DWI volumes with b=100-10,000 s/mm2. Processing of the DWI images was performed using TORTOISE software3 including DRBUDDI correction for geometric distortions4
DTI: Diffusion tensor5 fitting was performed using DIFFCALC tools with a weighted non-linear least squares algorithm and metric maps were generated including Trace and Fractional anisotropy (FA)6.
DKI: Diffusion and kurtosis tensors7 were fit using dke tools8 and maps were generated for the mean kurtosis (MK) and kurtois FA (KFA).
MAP-MRI: The mean apparent propagator9 was modeled using the DIFFCALC software package to generate maps for Non-Gaussianity (NG), return to the origin probability (RTOP) and propagator anisotropy (PA).
Detection and sensitivity were evaluated by voxelwise ANOVA with time following CCI as a factor using the randomise procedure of FSL and p-value maps were compared across metric maps along with ROI analysis.
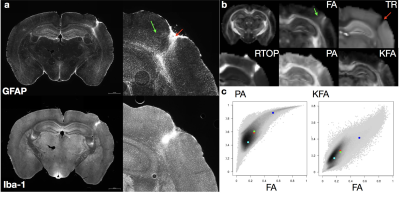
Specificity was evaluated by identification of histologic abnormalities and comparison with metric values in the same regions of the same brain which were evaluated qualitatively and using 2D histogram analysis.
Results
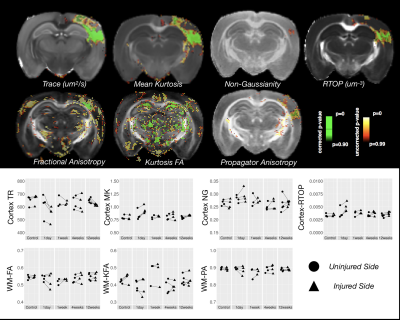
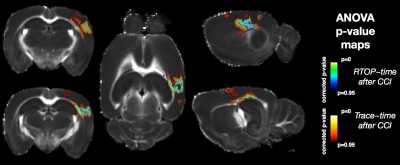
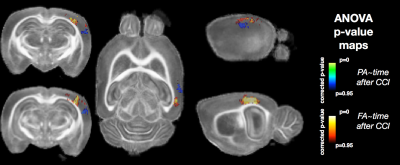
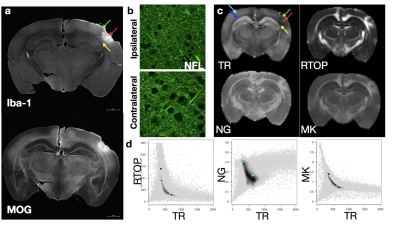
Voxelwise ANOVA p-value maps were found to show greater sensitivity of DTI compared with MAP-MRI scalars as the uncorrected and corrected p-value maps for TR and FA were more extensive than for DKI and MAP-MRI scalars (Figure 1). Two notable observations made by comparing the corrected p-value maps: 1 – the RTOP was observed to identify focal subregions within the affected area (Figure 2) and 2 – PA abnormalities were found to be non-overlapping with FA abnormalities (Figure 3). In regions of cellular damage 24 hours following CCI (Figure 4) TR was reduced in both the core and barrier regions, while increased MK, RTOP and NG were localized primarily to the barrier region. At 12 weeks after CCI (Figure 5) adjacent regions of perivascular gliosis and organized gliosis were observed to have distinct DTI, DKI and MAP-MRI profiles.Discussion and Conclusions
In general, the findings of this study suggest
that higher order models have lower sensitivity for detection of brain
abnormalities identified by DTI, but may detect new regions of abnormalities and
define subregions within larger regions of DTI abnormality. While individual
maps from higher order models demonstrated greater vulnerability to noise (especially
KFA and NG), there was greater stability in values across samples for some
measures (e.g. MK and RTOP) in the ROI analysis. While improved delineation of the barrier and
core regions of injured tissue appeared to be improved using RTOP and MK as compared
with TR, these metrics appear to be strongly related on 2D histogram analysis. Acknowledgements
We thank the intramural NICHD research program and the section for quantitative imaging and tissue science for scanning resources.References
1. Hutchinson EB, Schwerin SC, Avram AV, Juliano SL, Pierpaoli C. Diffusion MRI and the detection of alterations following traumatic brain injury. Journal of neuroscience research 2017.
2. Hulkower MB, Poliak DB, Rosenbaum SB, Zimmerman ME, Lipton ML. A decade of DTI in traumatic brain injury: 10 years and 100 articles later. AJNR American journal of neuroradiology 2013;34(11):2064-2074.
3. Pierpaoli C, Walker L, Irfanoglu M, Barnett A, Basser P, Chang L, Koay C, Pajevic S, Rohde G, Sarlls J. TORTOISE: an integrated software package for processing of diffusion MRI data. Book TORTOISE: an Integrated Software Package for Processing of Diffusion MRI Data (Editor ed^ eds) 2010;18:1597.
4. Irfanoglu MO, Modi P, Nayak A, Hutchinson EB, Sarlls J, Pierpaoli C. DR-BUDDI (Diffeomorphic Registration for Blip-Up blip-Down Diffusion Imaging) method for correcting echo planar imaging distortions. NeuroImage 2015;106:284-299.
5. Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophysical journal 1994;66(1):259-267.
6. Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Di Chiro G. Diffusion tensor MR imaging of the human brain. Radiology 1996;201(3):637-648.
7. Jensen JH, Helpern JA, Ramani A, Lu H, Kaczynski K. Diffusional kurtosis imaging: the quantification of non-gaussian water diffusion by means of magnetic resonance imaging. Magnetic resonance in medicine 2005;53(6):1432-1440.
8. Tabesh A, Jensen JH, Ardekani BA, Helpern JA. Estimation of tensors and tensor-derived measures in diffusional kurtosis imaging. Magnetic resonance in medicine 2011;65(3):823-836.
9. Özarslan E, Koay CG, Shepherd TM, Komlosh ME, İrfanoğlu MO, Pierpaoli C, Basser PJ. Mean apparent propagator (MAP) MRI: a novel diffusion imaging method for mapping tissue microstructure. NeuroImage 2013;78:16-32.
Figures