0119
Rapid MR Imaging of Ocular Movement using Shared K-Space Data for Radiotherapy Planning1Ophthalmology, Leiden University Medical Center, Leiden, Netherlands, 2Radiology, C.J. Gorter Center for High Field MRI, Leiden University Medical Center, Leiden, Netherlands
Synopsis
During ocular movement, the shape and location of the intra-orbital structures change. Outlining these changes will improve the accuracy of radiotherapy treatment planning for ocular tumors and the treatment of impaired ocular movement. The resolution of ocular MRI is, however, limited by acquisition time due to eye-motion. Therefore, an acquisition strategy in which outer k-space data is shared between gaze directions was developed and evaluated in 7 healthy and 2 myopic subjects. With this technique, the location of the orbital structures was determined for nine gaze directions in approximately one minute.
Introduction
During ocular movement, the shape and location of the intra-orbital structures, such as the optic nerve (ON) and extra-ocular muscles (EOMs), change. A rapid and accurate method to quantify these changes would be valuable for various ocular conditions. In radiotherapy of intraocular tumors, for example, the patient is asked to gaze in a specific direction to position the tumor in front of the radiation source. The planning is, however, performed in central gaze. This mismatch results in unnecessary damage to critical structures such as the ON.1,2 Furthermore, in highly myopic patients, the EOMs can hinder the rotation of the elongated eye, resulting in double vision.3,4
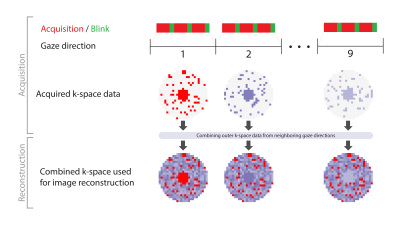
For a full map of the location of the intra-orbital structures as a function of gazing direction, at least 9 different directions need to be imaged. However, the spatial resolution of ocular MRI is highly limited by acquisition time, as patients can only keep their eyes still for a few seconds. A cued-blinking paradigm removed most of the blink-related motion artefacts,5,6 but total scan times are still limited as fatigue also results in motion artefacts. Therefore, a fast method was developed to image the complete eye and surrounding structures for nine gazing directions by sharing part of the acquired data between different gaze directions.
Methods
Both eyes of nine subjects (7 healthy subjects, 2 myopic patients) were scanned on a 7Tesla Phillips Achieva MRI using a 32-channel head-coil after giving informed consent. A 3D-MPRAGE scan (TI/TR/TE/FA:857ms/4.0ms/1.8ms/8º, shot interval:2.7s, SENSE:1.5x1.5, FOV:120x80x37mm3) with nine successive scans was modified to acquire the center of k-space together with a, randomly distributed, section of the outer k-space in each dynamic. The incomplete outer k-space data is completed with data from neighboring dynamics, figure 1. Two variants of this scan were acquired: variant 1 (resolution:(1.0mm)3, central k-space:30%, outer k-space density:17%, scan time:66s) and variant 2 (resolution:(0.9mm)3, central k-space:25%, outer k-space density:11%, scan time:75s). The subjects were presented nine fixation targets, one for each dynamic scan, from left to right. They were instructed to blink between each MPRAGE-shot.
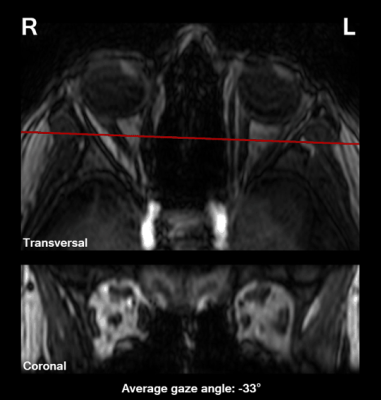
The ON and medial and lateral EOM were manually segmented in MIPAV on a coronal slice approximately 5 mm behind the globe. Gaze angle was measured on all dynamic scans. The ON location was determined for each gaze angle to evaluate the potential of this technique for radiotherapy planning, while EOM location and size were measured as these are key parameters in the evaluation of diplopia.
Results
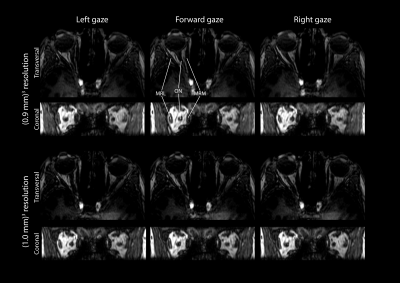
Although the higher-resolution scans were acquired with a high degree (75%) of shared data, this did not result in significant artifacts, figure 2,3. All subsequent evaluations were therefore performed on the higher-resolution scans.
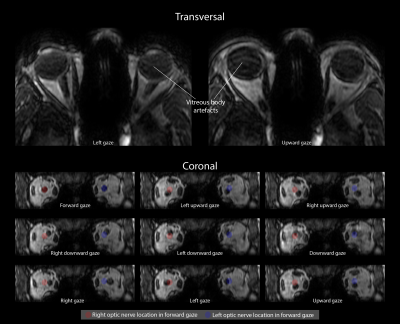
Execution of the fixation task differed per subject. Some subjects mainly fixated with their dominant eye, while others used their left eye for the left targets and the right eye for the right targets, causing gaze direction jumps, figure 3.
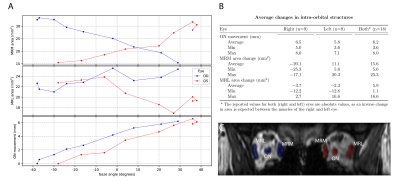
The average optic nerve movement between the far left and far right gaze was 6.2mm (min:3.6mm, max:8.0mm). The absolute change in muscle area was 15.6mm2 (min:5.6mm2, max:25.3mm2) for the medial EOM and 5.9mm2 (min:1.1mm2, max:16.6mm2) for the lateral EOM, figure 4.
Discussion
By sharing outer k-space data between different gaze directions, a full map of the ocular anatomy for 9 gaze directions was acquired without significant artifacts in approximately 1 minute. Although we currently only assessed the horizontal visual field, this can easily be extended to other directions, figure 5.
The presented single-slice analysis gives a first impression of the clinical potential of this technique for radiotherapy planning and diplopia evaluation. For the lateral EOMs, the small muscle area change was unexpected, but can be explained by the limitations of single slice analysis, as different parts of the muscles move into the slice during ocular movement.
Although the current resolution of (0.9mm)3 already provides relevant clinical information, a higher resolution would be desirable. Further tailoring of the acquisition scheme, for instance with higher undersampling and more sophisticated reconstruction techniques such as compressed sensing,7,8 would allow a higher-resolution acquisition within similar scan time. These techniques are needed to assess a greater variety of gaze directions, figure 5, as the lower degree of similarity between these images results in more severe artifacts originating from the shared outer k-space data.
Conclusion
By sharing outer k-space data across multiple gaze directions, the location and shape of the intra-orbital structures as a function of gaze direction can be mapped in approximately one minute. This has direct applications in the radiotherapy planning for ocular tumors and in the treatment planning for impaired ocular movement.Acknowledgements
The authors thank Maarten Versluis (Philips Healtcare, Best, the Netherlands) for assistance on the implementation of the protocols on the MRI scanner.References
1. Desjardins L, Lumbroso-Le Rouic L, Levy-Gabriel C, Cassoux N, Dendale R, Mazal A, et al. Treatment of uveal melanoma by accelerated proton beam. Developments in ophthalmology. 2012;49:41-57.
2. Bekkering GE, Rutjes AWS, Vlassov VV, Aebersold DM, von Bremen K, Jüni P, et al. The Effectiveness and Safety of Proton Radiation Therapy for Indications of the Eye. Strahlentherapie und Onkologie. 2009;185(4):211-21.
3. Aoki Y, Nishida Y, Hayashi O, Nakamura J, Oda S, Yamade S, et al. Magnetic resonance imaging measurements of extraocular muscle path shift and posterior eyeball prolapse from the muscle cone in acquired esotropia with high myopia. American journal of ophthalmology. 2003;136(3):482-9.
4. Krzizok TH, Schroeder BU. Measurement of recti eye muscle paths by magnetic resonance imaging in highly myopic and normal subjects. Investigative ophthalmology & visual science. 1999;40(11):2554-60.
5. Berkowitz BA, McDonald C, Ito Y, Tofts PS, Latif Z, Gross J. Measuring the human retinal oxygenation response to a hyperoxic challenge using MRI: eliminating blinking artifacts and demonstrating proof of concept. Magnetic resonance in medicine. 2001;46(2):412-6.
6. Beenakker JW, van Rijn GA, Luyten GP, Webb AG. High-resolution MRI of uveal melanoma using a microcoil phased array at 7 T. NMR in biomedicine. 2013;26(12):1864-9.
7. Candes EJ, Tao T. Near-Optimal Signal Recovery From Random Projections: Universal Encoding Strategies? IEEE Transactions on Information Theory. 2006;52(12):5406-25.
8. Donoho DL. Compressed sensing. IEEE Transactions on
Information Theory. 2006;52(4):1289-306.
Figures