0117
MRI-only Treatment Planning using Pseudo CT Generation from Deep Learning Approach1Department of Radiology, University of Wisconsin-Madison, Madison, WI, United States, 2Department of Human Oncology, University of Wisconsin-Madison, Madison, WI, United States
Synopsis
A MRI-only treatment planning pipeline, deepMTP, was constructed using a deep learning approach to generate continuously-valued pseudo CT images from MR images. A deep convolutional neural network was designed to identify tissue features in volumetric head MR images training with co-registered kVCT images. A set of 40 retrospective 3D T1-weighted head images was utilized to train the model, and evaluated in 10 clinical cases with brain metastases. Statistical analysis was used to compare dosimetric parameters of plans made with pseudo CT images generated from deepMTP to those made with kVCT based clinical treatment plan, where no significant difference was found.
Introduction
In recent years there have been many efforts to develop MRI-only treatment planning that avoid auxiliary CT for radiation therapy treatment planning (1). A key challenge for MRI-based treatment planning is the lack of a direct approach to obtain electron density for dose calculation. Given the importance of an accurate photon attenuation map to enable accurate dose calculation, the development of novel approaches to generate pseudo CTs from MRI is an actively-studied topic. While state-of-the-art image segmentation-based and atlas-based approaches can be of value, segmentation-based approaches typically require special MR sequences and atlas-based approaches may suffer when utilized in subjects with abnormal anatomy. Therefore there are still needs to develop versatile and robust approaches to generate pseudo CTs for treatment planning purpose. Deep learning utilizing convolutional neural networks (CNN) have recently been successfully implemented to generate pseudo CTs yielding excellent PET/MR attenuation correction performance(2). The purpose of this study was to develop and evaluate the feasibility of deep learning approaches for MRI-based treatment planning in brain tumor patients.Methods
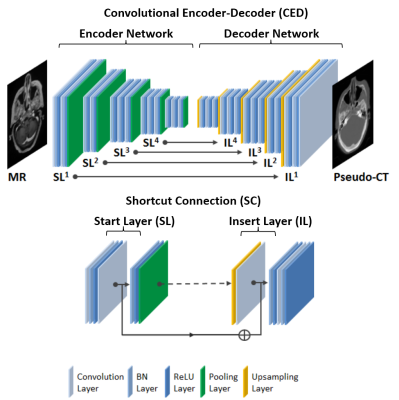
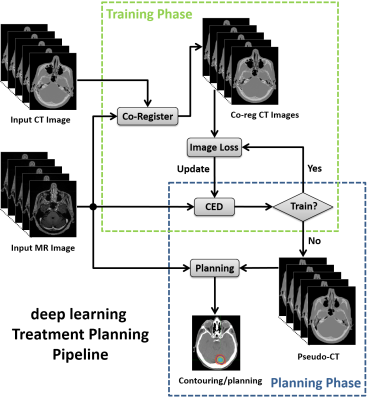
Specifically, we propose deepMTP which allows generation of a continuously-valued pseudo CT from a single MRI acquisition from clinical protocol using deep CNN model. Similar to study (2), we utilized the deep convolutional encoder-decoder (CED) network structure shown in Figure 1, which is capable of mapping pixel-wise image intensity from MRI to CT in multiple image scales. The encoder uses the same 13 convolutional layers from the VGG16 network(3). The decoder is applied directly after the encoder network and features a reversed VGG16 network structure with the max-pooling layers replaced by upsampling layers. As a result, a continuously-valued output is enabled. Additionally, shortcut connections (SC) were added between the encoder and decoder network to enhance the mapping performance of the encoder-decoder structure(4). The deepMTP procedure consisted of two independent phases for training retrospective MRI and co-registered CT data and for generating pseudo CTs using a fixed network in the treatment planning phase as shown in Figure 2. In current study, training was performed on 40 head images from who underwent both a T1-weighted post-contrast 3D MR scan and a non-contrast CT scan on the same day. Evaluation was performed on 10 randomly selected patients with brain metastases, treated with Fractionated Stereotactic Radiotherapy (FSRT) with acquired MR and kVCT scans. Dice coefficient was used to calculate the classification accuracy for soft tissue, bone, and air, where pseudo CT generated from deepMTP and the ground truth kVCT images were compared after discretization by HU threshold. A volumetric modulated arc therapy (VMAT) plans was generated with a 6 MV photon beam using 2-5 arcs with pseudo CT and kVCT (referred to as CTTP thereafter) respectively. Differences between deepMTP and CTTP in dose volume histogram (DVH), planning target volume (PTV), maximum dose, and V95 were calculated and compared.Results
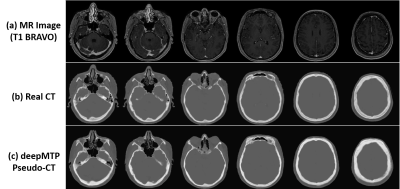
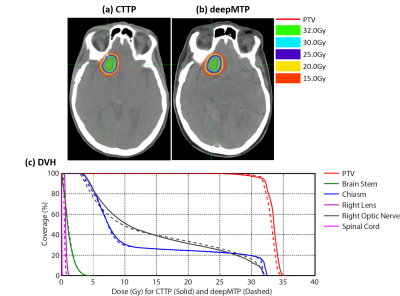
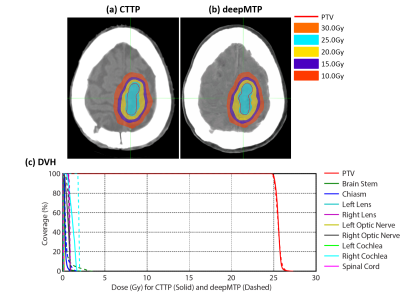
For the deepMTP procedure, the training phase required approximately 2.5 hours of computational time in our dataset, whereas generating a single pseudo CT image using the trained model and input MR images required roughly 1 minute. Evaluation of the Dice coefficient in 10 brain metastases cases comparing the output tissue mask from the pseudo CT (Figure 3) to the kVCT mask was high for air: 0.95 ± 0.01, soft tissue: 0.94 ± 0.02, and bone: 0.85 ± 0.02. The absolute percentage differences for deepMTP in comparison to CTTP was 0.24% ± 0.46% for PTV volume, 1.39% ± 1.31% for maximum dose and 0.27% ± 0.79% for V95. There was no significant difference between CTTP and deepMTP for PTV volume (p = 0.50), maximum dose (p = 0.83) as well as V95 (p = 0.19) in paired-sample Wilcoxon signed rank sum tests. Examples of two patients with right frontal brain tumor and a large superior brain tumor are demonstrated in Figure 4 and 5, respectively, showing the excellent agreement of isodose distribution and DVH between deepMTP and CTTP.Discussion
We have demonstrated that deep learning approaches applied to MR-based treatment planning in radiation therapy can produce comparable plans relative to CT-based methods. To the best of our knowledge, this is the first study to use and evaluate a deep learning approach for MR-based treatment planning. The further development of such approaches for MR-based treatment planning has potential value for providing accurate dose coverage and reducing treatment unrelated dose in radiation therapy, improving workflow for MRI-only treatment planning, combined with the improved soft tissue contrast and resolution of MR. Our study suggests that deep learning approaches such as deepMTP, as described herein, will have a substantial impact on future work in treatment planning in the brain and elsewhere the body.Acknowledgements
No acknowledgement found.References
1. Edmund JM, Nyholm T: A review of substitute CT generation for MRI-only radiation therapy. Radiat Oncol 2017; 12:28.
2. Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB: Deep Learning MR Imaging–based Attenuation Correction for PET/MR Imaging. Radiology 2017(September):170700.
3. Simonyan K, Zisserman A: Very Deep Convolutional Networks for Large-Scale Image Recognition. ArXiv e-prints 2014.
4. He K, Zhang X, Ren S, Sun J: Identity Mappings in Deep Residual Networks. ArXiv e-prints 2016.
Figures