0116
Translational Radiogenomics of Brain Tumors: From Lab-Invesigation to Clinical Application1Department of Neurosurgery, Medical Center Freiburg, Freiburg, Germany, 2Department of Nueroradiology, Medical Center Freibrurg, Freibrurg, Germany, 3Medical Center Freiburg, Freiburg, Germany
Synopsis
The interpretation of radiogenomic findings and its biological or clinical meaning is currently discussed and still unknown. Therefrom, a translational approach is aimed for integration of laboratory investigations and radiogenomic findings resulting in meaningful integration of radiogenomic discoveries. The presented study aimed to explore the molecular background of a MR-Spectroscopy imaging. In a stepwise approach findings were validated on tissue samples and finally taken into glioma cell models.
INTRODUCTION
Radiogenomics is a fast expanding research area, which combines radiology, molecular data and omics science. This analytic approach allows an automated and sophisticated method for clinical diagnostics. Nevertheless, the interpretation of radiogenomic findings and its biological or clinical meaning is currently discussed and still unknown. Therefrom, a translational approach is aimed for integration of laboratory investigations and radiogenomic findings resulting in meaningful integration of radiogenomic discoveries. The presented study aimed to explore the molecular background of a MR-Spectroscopy-Radiogenomic study1. In a stepwise approach findings were validated on tissue samples and finally taken into glioma cell models (Figure 1).METHODS
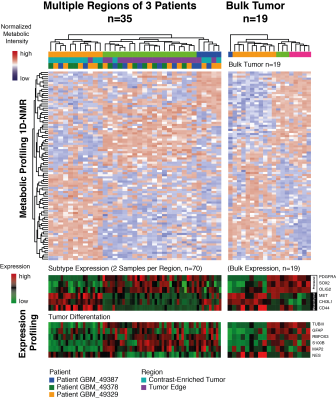
In a recent study, the connection between creatine metabolism and the proneural transcriptional subclass of glioma was uncovered1,2. The first aim was to explore the metabolic distribution and heterogeneity within the same tumors. Multiple biopsies (total n=138) were obtained from three de-novo glioblastoma multiforme patients and underwent a metabolic and transcriptional profiling. Computation of the metabolic-transcriptional network confirmed the association of creatine and the proneural gene expression. In a second step, patient derived glioblastoma stem-like cells (GSC) were cultured in either hypoxia, creatine- supplemented or combined conditions for 24h and 48h. Extensive metabolic, proteomic and transcriptional profiling was performed. These laboratory investigations were taken into consideration and validated on a cohort of 102 patients, who underwent MR-Spectroscopic imaging.RESULTS
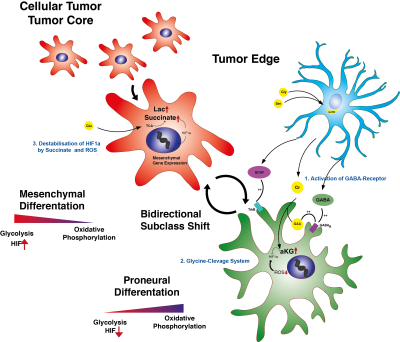
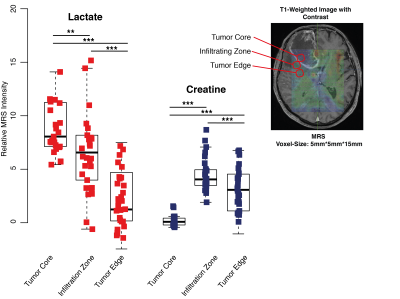
A metabolic landscape of multiple biopsies was explored marked by varying stages of hypoxia and creatine enrichment with a strong correlation to proneural transcriptional pattern. Therefrom, we aimed to explore the molecular background of creatine in a glioma cell-model. This environmental cell-model showed a synergistic effect of up-regulating the GABA signaling and a simultaneous inhibition of HIF signaling in a creatine-supplemented environment. This affected the transcriptional landscape with a shift toward a proneural subtype expression followed by a slowing of cell migration and proliferation. These findings were verified by additional HIF and GABA inhibition, which partially rescued the proneural shift. In a heterogeneous tumor, different stages of hypoxia and environmental creatine enrichment were observed, which required a permanent adaptation of the tumor to varying environmental conditions. Additionally, creatine enrichment was mainly observed in the tumor leading edge and infiltration regions. Next we examined this hypothesis in 102 patients with MR-Spectroscopic imaging. Patients with creatine enrichment in the non-resected FLAIR hyperintense regions showed a significantly lower progression-free survival (HR 0.53 95%CL 0.3-0.68, p<0.01).DISCUSSION
This study showed a new concept for radiogenomic explorations and interpretation of radiogenimic findings. By a stepwise approach, we uncovered the metabolic heterogeneity of glioblastoma and confirmed the important role of the metabolite creatine. The laboratory investigations were translated to MRS imaging and implemented creatine as a new marker for tumor invasion. Translational aspects of radiogenomic can improve its biological and clinical meaning and advance future tumor-imaging.Acknowledgements
No acknowledgement found.References
Heiland, D. H. et al. Integrative Network-based Analysis of Magnetic Resonance Spectroscopy and Genome Wide Expression in Glioblastoma multiforme. Sci. Rep. 6, 29052 (2016).
Heiland, D. H. et al. The integrative metabolomic-transcriptomic glioblastome multiforme landscape of. Oncotarget (2017).
Figures