0060
A 2D multi-shot inversion recovery EPI (MS-IR-EPI) sequence for high spatial resolution T1-mapping at 7TRosa Sanchez Panchuelo1, Robert Turner1,2,3, Olivier Mougin1, and Susan Francis1
1Sir Peter Mansfield Imaging Centre, University of Nottingham, Nottingham, United Kingdom, 2Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 3University of Amsterdam, Amsterdam, Netherlands
Synopsis
The present study uses an efficient 2D, multi-shot, inversion-recovery EPI (MS-IR-EPI) acquisition that combines separately excited k-space segments after each inversion pulse together with steps in slice ordering to generate T1-maps with high SNR per unit time. We show that although the inversion times systematically vary across slices, consistent T1-maps can be generated across the whole brain. Such T1-maps provide high spatial resolution and SNR, with little image distortion and can be collected in a short acquisition time.
Introduction
Cortical parcellation for unambiguous in-vivo identification of distinct cortical areas has been performed using high resolution MRI that provides some details of myeloarchitecture [1-3]. Quantitative T1 maps provide evidence of heavily myelinated layers [4] and can easily be obtained with high spatial resolution at 7T [5]. However, the low flip-angle 3D FLASH-type sequences which are commonly used for T1 mapping have poorer SNR per unit time compared with techniques such as EPI and GRASE that use high flip angles [6]. Further, when inversion pulses are efficiently included in 3D sequences, the point spread function (PSF) becomes broad, because the acquisition window is wide compared with T1. An alternative approach is to use 2D inversion-recovery EPI which has high SNR per unit time and a short echo train length compared with T1 [7]. However, if each slice in a multislice brain volume is required to have the same inversion time, the scan time becomes long, especially if segmentation is added to improve spatial resolution, because the longitudinal magnetization must recover after each inversion pulse. Removing these requirements can greatly accelerate data acquisition, but entails the computation of T1-maps for each slice using different inversion times. Here we develop an efficient 2D, multi-shot, inversion-recovery EPI (MS-IR-EPI) acquisition combining separately excited k-space segments after each inversion pulse and shifted slice ordering to provide high SNR per unit time.Methods
A 2D MS-IR-EPI sequence 0.5x0.5mm2 in-plane resolution (TE=20ms, TR=3.2s, EPI-factor=17, 12 shots, field-of-view (FOV)=180(AP)x154(RL), SENSE=1.5(RL), 38.4s acquisition time) was evaluated at 7T for T1-mapping. Data sets consisting of 24 axial slices (1.5mm slice thickness) were acquired with varying slice ordering schemes, thus providing a number of equally spaced inversion times for each slice (Fig.1). Data was acquired at a number of slice offsets=[0, 4, 5, 6, 8, 10, 12, 15, 16, 18, 20] and used to reconstruct T1-maps based on 2, 3, 4, 5 or 6 slice offsets and thus range of inversion times (TI). Data sets were acquired with and without fat suppression. In addition, a single slice dataset was acquired using standard IR-EPI at 10 TIs=[100, 200, 400, 600, 900, 1100, 1300, 1600, 2100, 3100], TR=5s. To compute T1-maps, the polarity of the modulus data was corrected (based on phase) and fitted for T1 and S0: S(TI) = S0[1-2exp(−TI/T1)+exp(-TR/T1)]. For the MS-IR-EPI, Monte Carlo simulations were performed to evaluate the accuracy and variance of the fitted T1s with different numbers of TIs and different level of noise (1024 repeats). High spatial resolution (0.5 mm isotropic) data were collected on two subjects using a MS-IR-EPI (48 slices, 4 TIs/slice offsets each with 3 averages, 461s total acquisition time) and MP2RAGE sequence (TR/TE=14/6.4ms, TI1/TI2=680/2080ms, TRshot=3.5s, SENSE=2 (RL), 724s acquisition time) with matched FOV but 100 slices coverage. A PSIR-reconstruction [8] of the MP2RAGE data was used to generate images proportional to T1.Results and discussion
T1-maps from the MS-IR-EPI sequence were homogeneous across slices, despite each slice being fit with different inversion times (Fig.2A), and were independent of the number of TIs when more than 2 inversion times were used in the fit (Fig.2B). Fitted T1-values were shorter than those obtained with a single slice standard IR-EPI sequence (Fig.2B). This difference was reduced when collecting data with no fat suppression, suggesting this discrepancy is likely due to magnetization transfer effects [9]. Future work will explore the use of different fat suppression techniques while reducing the impact on fitted T1-values. A Monte Carlo simulation showed that the accuracy of the fitted T1 is independent of the number of TIs used in the fit whilst the standard deviation decreases as the number of TIs increases (Fig.3A), and as the SNR increases (data not shown). Although the fitted T1 remains relative constant across slices (Fig.2B) there is variation across slices when using 2 and 3 TIs for WM (Fig.3B) and 2 TIs for GM (Fig.3C). Figures 4 and 5 compare the 0.5mm isotropic acquisition T1-maps generated from the MS-IR-EPI (no fat suppression) with MP2RAGE processed images. Both methods yield similar SNR, but note that while the MS-IR-EPI is based on a true T1-fit, MP2RAGE is currently proportional to T1, and future work will generate a look-up table to estimate T1 [5]. Figure 5 shows that the 2D MS-IR-EPI T1-maps have minimal image distortion (B) and are sharper than 3D MP2RAGE maps (Fig.5C).Conclusion
High quality T1-maps can be generated using a high resolution 2D MS-IR-EPI sequence. Future work will combine the MS-IR-EPI scheme with simultaneous multi-slice excitation (SMS) to achieve whole brain coverage in short acquisition times.Acknowledgements
This work was funded by a MRC grant MR/M022722/1 and a Leverhulme fellowship.References
[1] Geyer S, Weiss M, Reimann et al. Microstructural Parcellation of the Human Cerebral Cortex – From Brodmann's Post-Mortem Map to in vivo Mapping with High-Field Magnetic Resonance Imaging Front Hum Neurosci. 2011;18:5-19. [2] Sánchez-Panchuelo RM et al, Francis S, Schluppeck D & Bowtell R. Correspondence of human visual areas identified using functional and anatomical MRI in vivo at 7 T. J Magn Reson Imaging. 2012;35(2):287-99 [3] Glasser MF, Coalson TF, Robinson EC et al. Nature. A multi-modal parcellation of human cerebral cortex. 2016;536(7615):171-178. [4] Stüber C, Morawski M, Schaeffer A et al. Myelin and iron concentration in the human brain: A quantitative study of MRI contrast. Neuroimage. 2014;93(Pt 1):95-106 [5] Marques J, Kober T, Krueger G et al. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage. 2010;49(2):1271-81. [6] Trampel R, Reimer E, Hueber L, et al. Anatomical brain imaging at 7T using two-dimensional GRASE. Magn Reson Med. 2014;72(5):1291-301 [7] Mansfield P, Guilfoyle DN, Ordidge RJ and Coupland RE. Measurement of T1 by echo-planar imaging and the construction of computer-generated images. Phys Med Biol. 1986;31(2):113-24. [8] Mougin et al. Abdel-Fahim R, Denin R et al. Imaging grey matter with concomitant null point imaging from the phase sensitive inversion recovery sequence. MRM. 2016;76(5):1512-1516. [9]. Wright ), Mougin O, Totman JJ, et al. Water proton T1 measurements in brain tissue at 7, 3, and 1.5T using IR-EPI, IR-TSE, and MPRAGE: results and optimization. MAGMA. 2008;21:121.Figures
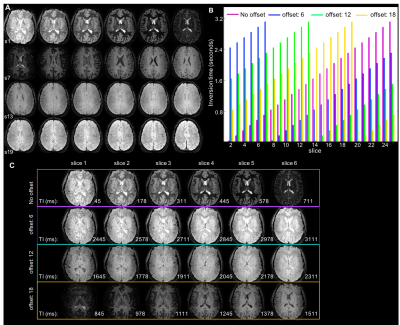
(A) Example 2D multi-shot
inversion recovery EPI. Twenty-four slices were acquired in ascending order.
The different contrast across slices reflects the different TI (equally
distributed between 0 and 3.2s). (B) To generate T1-maps, the slice acquisition
order was varied by adding a slice offset to the default ascending order. The distribution
of inversion times for each of 24 slices is shown for 4 acquisitions with slice
offsets=[0,6,12,18], each colour representing a different offset. (C) First 6
slices for each of the 4 acquisition schemes. Note that for each slice the 4
TIs are equally spaced at 800 ms intervals.
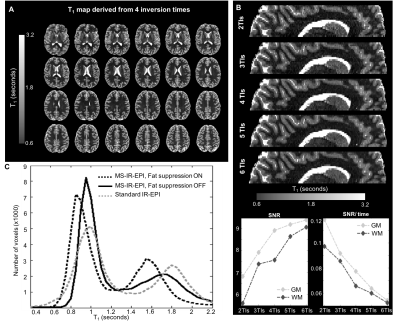
(A) T1-maps generated
using the acquisition described in Figure 1. (B) Sagittal reconstructions of the
axial T1-maps derived using 2, 3, 4, 5 and 6 equally spaced TIs,
with SNR and SNR/time plot against number of TIs (bottom). Fitted T1-values
are independent of the number of TIs and are homogeneous across slices. (C) T1-histograms
of a single slice (map derived using 6 TIs) showing the effect of fat
suppression (dashed-black line), which shortens the fitted T1-values.
When no fat suppression is applied the T1-values (solid-black line)
become closer to the standard IR-EPI mapping for a single slice acquisition
(dashed-gray line).
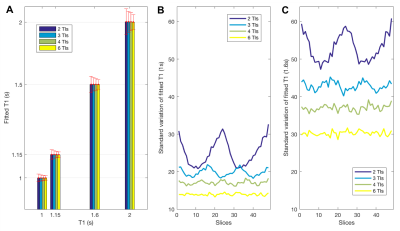
Monte Carlo simulations showing
the effect of number of TIs (2,3,4,6) on fitted parameters. (A) Simulations
were performed for target T1=(1, 1.15, 1.6, 2 s) and SNR=25 (representative
of the 0.5 mm isotopic data) and 48 slices. Random noise was added to the simulated
data which was fit as for the experimental data to obtain 1024 fitted T1
data sets. (A) The fitted T1 (mean across slices) remains constant,
but the standard deviation (error bars) decreases with increasing number of TIs.
(B) Standard deviation across each of 48 slices for fitted T1 of 1s
(~WM) and (C) 1.6s (~GM).
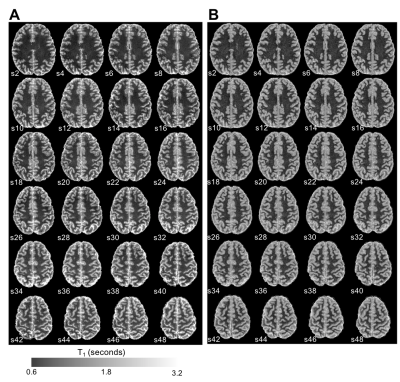
(A) High resolution (0.5 mm isotropic) T1-maps generated from 2D MS-IR-EPI acquisitions with 4 TIs/slice offsets. Three
averages were acquired for each acquisition to improve SNR (460s total
acquisition time). Data shown for each
even slice. (B) PSIR reconstruction (inverted) of the MP2RAGE acquisition (724s)
shown for the same set of slices as in (A).
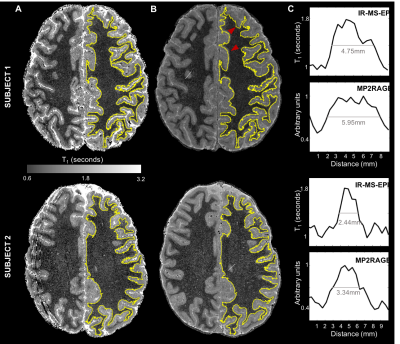
Example slice from (A) a T1-map
generated with 2D MS-IR-EPI and (B) MP2RAGE processed image. GM/WM boundaries
(yellow line) were generated from the EPI maps and overlaid onto the MP2RAGE
space, showing that geometric distortions (pointed by the red arrowheads) are
minimal. C. Profiles along GM/WM boundaries indicated by the white line (in
panel B) are shown for both IR-MS-EPI (top) and MP2RAGE (bottom), showing that
the MP2RAGE data seems to have a broader point-spread function.