0041
Bundle-Wise Deep Tracker: Learning to track bundle-specific streamline paths1University of Sherbrooke, Sherbrooke, QC, Canada
Synopsis
We propose a novel bundle-wise tracking algorithm based on deep learning and recurrent neural networks. This allows bundle-specific features to be learned directly from the diffusion signal without the need to reconstruct a fiber orientation distribution. With a high amount of examples, the proposed method improves classic algorithms for several quantitative measures such as tracking efficiency, number of valid streamlines, and volume coverage.
Introduction
Recently, a few models based on deep learning1,2,3 were recently proposed to do whole-brain tracking, and suggests that while learning methods can achieve results comparable to classic tractography algorithms, they are still prone to make the same mistakes. We choose to leverage deep learning methods and instead explore bundle-wise tractography. However, even if tracking is restrained to a single bundle, the diffusion signal can still represent other bundles, and tracking is prone to follow those pathways. In such a case, machine learning methods are extremely relevant, as they learn discriminant features in order to make predictions, i.e. which features of the diffusion signal explain the bundle. In this work, we use training streamlines generated by Bundle-Specific Tracking (BST)4, which computes bundle-wise directional priors in MNI space using reference subjects in order to enhance fiber ODFs5. We evaluate algorithms using a neuroanatomist “virtual dissection” to automatically segment valid streamlines.Dataset
Diffusion-weighted images (DWI) were collected from 37 subjects in the BIL&GIN database (2 acquisitions of 2x21 directions, b=1000s/mm2, 2mm isotropic)6. Subjects were randomly divided into training (20), validation (5) and test sets (12).Methods
Building on the architecture of Learn-to-Track1, we propose the Bundle-Wise Deep Tracker (BWDT), a Recurrent Neural Network (RNN) trained to predict tracking directions using bundle-wise input data. GRU7 was chosen as the RNN implementation, using a hidden layer of size 512 with layer normalization8. Optimization was done with the Adam9 optimizer, an initial learning rate of 0.0001 and gradient clipping of norm 5. During training, an L2 reconstruction error was minimized between normalized predictions and targets, with early stopping of 10 epochs. Models were regularized using a variant of Dropout called Zoneout10 with a drop probability of 0.3, and random 3D gaussian noise was added to the streamline coordinates (0.03mm) across all batches.
Reference streamlines were generated using particle filtered probabilistic BST4,11, segmented by a neuroanatomy expert and resampled to a constant step size of 1.0mm. Model inputs were interpolated at reference positions from raw DWIs fitted with spherical harmonics of order 812.
Tracking
For each method and test subject, tracking was done with 5 seeds per voxel in the white matter (WM) - gray matter (GM) interface, using a dilated WM tracking mask. Baseline methods use a step size of 0.2mm, while our method uses a step size of 1.0mm (given the recurrent nature of the model, smaller step sizes tend to worsen performance).Evaluation
Three algorithms were evaluated as baseline results:
- Deterministic (DET) tracking13
- DET BST4
- Probabilistic (PROB) BST4 with particle filtering11
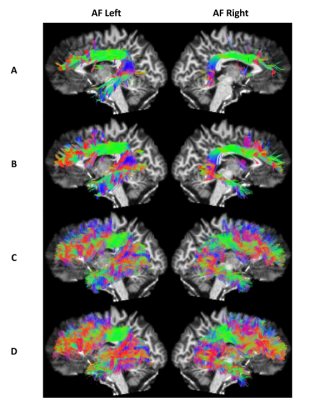
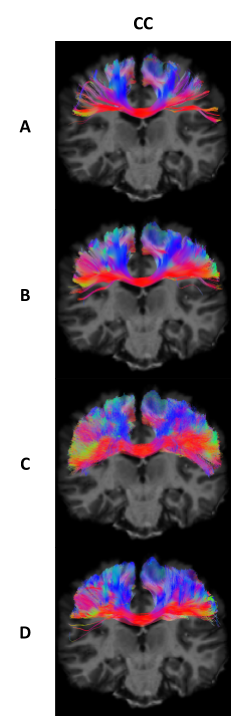
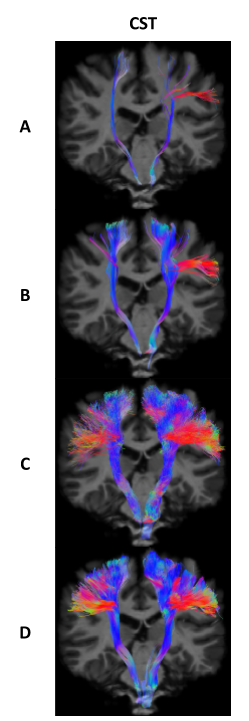
All methods tracked five bundles of the randomly chosen 12 test subjects (Arcuate Fasciculus left/right, Corpus Callosum, Corticospinal Tract left/right). After initial tracking, automatic segmentation was done using the same procedure as in BST4 to extract valid streamlines. Across all test subjects, 5 metrics were computed in template space: number of streamlines, volume coverage, valid streamline ratio, efficiency ratio (number of valid tracking steps over total number of tracking steps), and weighted dice score.
Results & Discussion
A model was trained for each bundle using an Nvidia TITAN Xp GPU, which took between 6 and 12 hours per model (depending on the number of streamlines in a bundle’s training set). Results are detailed in Table 1.
When enough training data is provided (AF Left/Right, CC), BWDT provides a sharp increase in valid and efficiency ratios. In other words, less streamlines are rejected, and rejected streamlines are generally shorter, which leads to faster tracking when generating the same number of valid streamlines. Note that even though BWDT is a deterministic model, its volume coverage is better or comparable to a state-of-the-art probabilistic tracking with particle filtering, and is much better than other deterministic methods, as seen in Figures 1, 2 and 3. Visually, BWDT also generates a much smoother streamlines than probabilistic tracking while keeping a high volume coverage. It also runs on a GPU implementation instead of CPU, and can track 200,000 streamlines in about 3 minutes, compared to 2 hours for PROB and 4 hours for DET (all on CPU).
Compared to other methods, BWDT is much easier to use, as it uses only DWI as input and does not need standard pre-processing steps to be robust to noise and motion artefacts. It also does not require registration as in BST4.
Conclusion
The proposed BWDT model outperforms other deterministic methods, and is generally comparable to state-of-the-art bundle-specific probabilistic methods. Given computational efficiency and GPU implementation of BWDT, tracking times are reduced by an order of magnitude compared to other methods.Acknowledgements
No acknowledgement found.References
[1] Poulin, P., Cote, M.A., Houde, J.C., Petit, L., Neher, P.F., Maier-Hein, K.H., Larochelle, H. and Descoteaux, M., 2017. Learn to Track: Deep Learning for Tractography, MICCAI 2017, Lecture Notes in Computer Science, vol 10433. Springer, Cham.
[2] Neher, P.F., Cote, M.A., Houde, J.C., Descoteaux, M. and Maier-Hein, K.H., 2017. Fiber tractography using machine learning. bioRxiv, p.104190.
[3] Koppers, S. and Merhof, D., 2016, October. Direct estimation of fiber orientations using deep learning in diffusion imaging. In International Workshop on Machine Learning in Medical Imaging (pp. 53-60). Springer International Publishing.
[4] Rheault, F., St-Onge, E., Chenot Q., Petit L., Descoteaux M., 2017. Bundle-Specific Tractography. Computational Diffusion MRI, MICCAI Workshop. Springer, CDMRI'17. - (Accepted)
[5] Tournier, J.D., Calamante, F. and Connelly, A., 2007. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage, 35(4), pp.1459-1472.
[6] Mazoyer, B., Mellet, E., Perchey, G., Zago, L., Crivello, F., Jobard, G., Delcroix, N., Vigneau, M., Leroux, G., Petit, L. and Joliot, M., 2016. BIL&GIN: a neuroimaging, cognitive, behavioral, and genetic database for the study of human brain lateralization. Neuroimage, 124, pp.1225-1231.
[7] Chung, J., Gulcehre, C., Cho, K. and Bengio, Y., 2014. Empirical evaluation of gated recurrent neural networks on sequence modeling. NIPS Deep Learning Workshop, 2014.
[8] Ba, J.L., Kiros, J.R. and Hinton, G.E., 2016. Layer normalization. arXiv preprint arXiv:1607.06450.
[9] Kingma, D. and Ba, J., 2014. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980.
[10] Krueger, D., Maharaj, T., Kramár, J., Pezeshki, M., Ballas, N., Ke, N.R., Goyal, A., Bengio, Y., Larochelle, H., Courville, A. and Pal, C., 2016. Zoneout: Regularizing rnns by randomly preserving hidden activations. arXiv preprint arXiv:1606.01305.
[11] Girard, G., Whittingstall, K., Deriche, R. and Descoteaux, M., 2014. Towards quantitative connectivity analysis: reducing tractography biases. Neuroimage, 98, pp.266-278.
[12] Descoteaux, M., Angelino, E., Fitzgibbons, S. and Deriche, R., 2006. Apparent diffusion coefficients from high angular resolution diffusion imaging: Estimation and applications. Magnetic Resonance in Medicine, 56(2), pp.395-410.
[13] Garyfallidis, E., Brett, M., Amirbekian, B., Rokem, A., Van Der Walt, S., Descoteaux, M., Nimmo-Smith, I. and Contributors, D., 2014. Dipy, a library for the analysis of diffusion MRI data. Frontiers in neuroinformatics, 8.
Figures
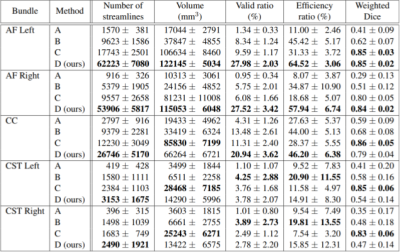
Table 1: Evaluation metrics on the test set, with means and standard deviations computed across 12 subjects.
A: DET; B: DET-BST; C: PROB-BST + PFT; D: BWDT