0038
Reducing false positives in tractography with microstructural and anatomical priors1Computer Science department, University of Verona, Verona, Italy, 2Signal Processing Laboratory 5 (LTS5), Ecole Polytechnique Federale de Lausanne, Lausanne, Switzerland, 3Radiology department, University Hospital Center (CHUV) and University of Lausanne (UNIL), Lausanne, Switzerland, 4Sherbrooke Connectivity Imaging Laboratory (SCIL), University of Sherbrooke, Sherbrooke, QC, Canada, 5Department of Nuclear Medicine and Radiobiology, Faculty of Medicine and Health Science, Sherbrooke Molecular Imaging Center, Sherbrooke, QC, Canada
Synopsis
Tractography has proven particularly effective for studying noninvasively the neuronal architecture of the brain but recent studies have showed that the high incidence of false positives can significantly bias any connectivity analysis. We present a novel processing framework that can dramatically reduce these false positives, i.e. improving specificity, without affecting the sensitivity, by considering two very basic observations about white-matter anatomy. Our results may have profound implications for the use of tractography to study brain connectivity.
Context and motivation
Over the years, tractography has proven particularly effective for studying noninvasively the neuronal architecture of the brain. However, recent studies have challenged its use for connectivity analyses, notably showing that the presence of false positive connections in the reconstructions can significantly bias the topological properties of the estimated brain networks1,2,3. In this study, we present a novel processing framework that can dramatically reduce the incidence of these false positives, i.e. improving specificity, without affecting the true ones, i.e. sensitivity. Our formulation combines basic prior knowledge about brain anatomy with group-sparsity regularization into an efficient convex optimization problem. In fact, tractography algorithms usually exploit only the directional information estimated with diffusion MRI: we speculate that this information is not enough and we advocate the need for additional data to help tractography producing more accurate reconstructions. We evaluated quantitatively the effectiveness of our formulation on different tractography reconstructions using a realistic digital phantom with known ground-truth.
Methods
COMMIT. Convex Optimization Modeling for Microstructure Informed Tractography4,5 (COMMIT) is a framework to complement tractography with additional microstructural information about the neuronal tissue. The originality of this solution lies in the possibility to express tractography and tissue microstructure in a unified formulation using convex optimization. The observation model is $$$\boldsymbol{y} = \boldsymbol{A} \boldsymbol{x} + \eta$$$, where $$$\boldsymbol{A}$$$ is the linear operator implementing the specific multi-compartment model adopted to characterize the neuronal tissue, $$$\boldsymbol{x}$$$ are the contributions of all compartments of the model to explain the DWI data $$$\boldsymbol{y}$$$ and $$$\eta$$$ accounts for noise and modeling errors. The system is solved as a least-squares problem: $$$\operatorname{argmin}_{\boldsymbol{x} \ge 0} \; || \boldsymbol{A} \boldsymbol{x} - \boldsymbol{y} ||_2^2$$$.
Adding anatomical priors. We extended COMMIT by considering two very basic observations about WM anatomy: (i) streamlines are not "just lines" but represent neuronal fibers, and (ii) they are organized in bundles. To enforce the first assumption, we implemented a simple model in $$$\boldsymbol{A}$$$ that assigns a contribution, i.e. volume or cross-sectional area, to each streamline proportionally to its length inside each voxel. Then, the total amount of streamlines that traverse a voxel must sum up to the intra-axonal volume fraction estimated in that voxel, which can be estimated e.g. using standard models like NODDI6 or SMT7. To implement the second prior, we clustered all streamlines connecting the same pair of ROIs into groups and then added a regularization term to try explaining the data using the minimum number of bundles; this is achieved by solving the problem using the group lasso: $$\operatorname{argmin}_{\boldsymbol{x} \ge 0} \; || \boldsymbol{A} \boldsymbol{x} - \boldsymbol{y} ||_2^2 \; + \; \lambda \, \sum_{g \in \mathcal{G}} || \boldsymbol{x}^{(g)} ||_2$$ where $$$\mathcal{G}$$$ represents a partition of the streamlines into groups.
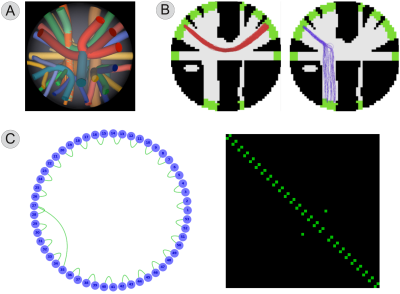
Experimental settings. We tested different tractography algorithms on the ISBI Challenge 2013 dataset, which consists of 27 fiber bundles mimicking the major WM tracts of the brain (Fig. 1A). The DWI signal was simulated using Phantomas8 according to the CHARMED model9, 64 directions, b-value=3000 s/mm2, and Rician noise (SNR=30). The ground-truth connectivity of the dataset is known (Fig. 1C), so we could quantitatively evaluate the reconstructions in terms of True Positive (TP) and False Positive (FP) bundles (Fig. 1B). Due to space limitations, we report only the results for deterministic tracking and the following parameters: 1 million fibers, angle-threshold $$$45^{\circ}$$$; results were consistent for any combination of parameters and tracking method.
Results
Fig. 2A reports the number of True Positives (TP) and False Positives (FP) in the raw tractogram. We can easily see that the tracking algorithm was able to reconstruct all 27 TP, i.e. high sensitivity, but at the price of recovering 235 FP, i.e. very low specificity. This result agrees with previous literature1,2,3. On the other hand, Fig. 2B clearly shows that using COMMIT the number of FP can be dramatically reduced (from 235 to 18) without affecting the sensitivity (the tractogram still contains all 27 TP).
Discussion and Conclusion
Recent studies have shown that current tractography algorithms are quite good at reconstructing the major WM bundles, i.e. high sensitivity, but at the price of also recovering a high incidence of false positive ones, i.e. low specificity. In particular, those false positive streamlines can dramatically bias any subsequent analysis, as non-existent structures are mixed with real ones. In this work, we showed that adding basic prior information on the organization of the neuronal tissue can help tractography in dramatically improving the quality of reconstructions. Our results represent an important step forward to improve the accuracy of tractography and may have profound implications for the use of tractography to study brain connectivity.
Acknowledgements
This work is supported by the Center for Biomedical Imaging (CIBM) of the Geneva-Lausanne Universities and the EPFL, as well as the foundations Leenaards and Louis-Jeantet.References
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA,Pierpaoli C. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci USA. 111(46):16574-9 (2014)
- Zalesky A, Fornito A, Cocchi L, Gollo LL, van den Heuvel MP, Breakspear M. Connectome sensitivity or specificity: which is more important? Neuroimage. 142:407-420 (2016)
- Maier-Hein KH et al. The challenge of mapping the human connectomebased on diffusion tractography. Nature Communications. 8:1349 (2017)
- Daducci A, Dal Palú A, Lemkaddem A and Thiran JP. A convex optimization framework for global tractography. In Proc. IEEE ISBI, 524–7 (2013)
- Daducci A, Dal Palú A, Lemkaddem A and Thiran JP. COMMIT: Convex Optimization Modeling for Microstructure Informed Tractography. IEEE Trans Med Imaging. 33(1):246–57 (2014)
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 61(4):1000-16 (2012)
- Kaden E, Kelm ND, Carson RP, Does MD, Alexander DC. Multi-compartment microscopic diffusion imaging. Neuroimage. 139:346-359 (2016)
- Caruyer E, Daducci A, Descoteaux M, Houde J-C, Thiran J-P, Verma R. Phantomas: a flexible software library to simulate diffusion MR phantoms. In Proc. of ISMRM (2014)
- Assaf Y, Basser PJ. Composite hindered and restricted modelof diffusion (CHARMED) MR imaging of the human brain. NeuroImage. 27(1):48–58 (2005)
Figures
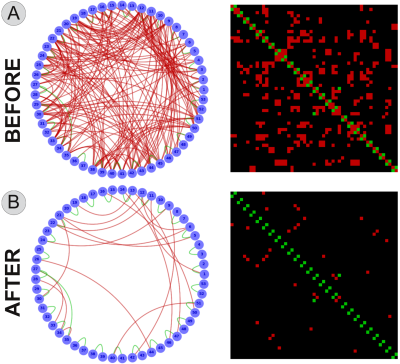
Sensitivity and specificity of a tractogram before (A) and after (B) applying the proposed method. In both plots, we identify the true positives in "green" and the false positives in "red". Please note that before applying COMMIT the raw tractogram contained TP=27 and FP=235 (the true positives are barely visible) whereas after COMMIT the tractogram still contained all the 27 TP but the FP were dramatically decreased from 235 to 18. Results hold also for other combinations of tracking method and parameters.