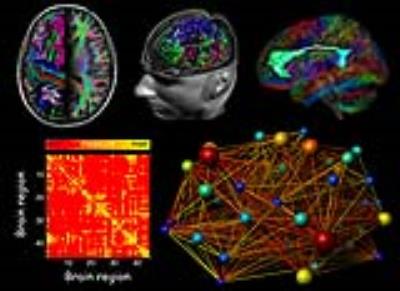
Measuring Connectivity with Diffusion MRI
1Image Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands
Synopsis
I will present the pipeline that is used for computing estimates of structural brain connectivity as obtained with diffusion tractography. Several methodological considerations will be discussed.
Computing estimates of structural connectivity with diffusion MRI consists of several steps in a lengthy pipeline (see figure) [Sporns, 2005]. In this presentation, I will discuss some of the challenges that one typically encounters, such as:
1) Which diffusion MRI approach (diffusion tensor imaging, spherical deconvolution, …)?
2) Which tractography algorithm?
3) Which definition of “connections” (edges)?
4) Which brain parcellation atlas (nodes)?
5) Which statistical approach (multiple comparison correction)?
6) Visualization?
Acknowledgements
The research of Alexander Leemans is supported by VIDI Grant 639.072.411 from the Netherlands Organisation for Scientific Research (NWO).References
Sporns O, Tononi G, Kotter R (2005): The human connectome: A structural description of the human brain. PLoS Comput Biol 1:e42.