5665
Deep learning: Utilizing the potential in data bases to predict individual outcome in acute stroke1Center of Functionally Integrative Neuroscience and MINDLab, Institute of Clinical Medicine, Aarhus University, Aarhus C, Denmark, 2Combat Stroke, Aarhus C, Denmark
Synopsis
Acute ischemic stroke is a major disease and one of the leading causes of adult death and disability. Brain tissue infarcts permanently within hours after onset and rapid reperfusion treatment is therefore of utmost importance. Current methods to predict the tissue outcome are too simplistic. In this project a more advanced approach using deep neural networks to utilize the information from previous patient developments was established and compared to current state-of-the-art. The predictions from the deep neural networks were showed to be superior to the state-of-the-art method improving prediction accuracy and hence leading to better decision support.
Introduction:
Each year, 15 million people worldwide suffers an acute ischemic stroke. Approximately one third of these incidents leads to permanent disability and another third leads to death, making acute ischemic stroke the third leading cause of disability. Brain tissue infarcts rapidly (two million neurons die every minute a major artery is occluded1) and rapid reperfusion through endovascular and/or thrombolytic treatment is therefore of utmost importance to ensure optimal outcome. The therapeutic strategy today is based on images of the brain obtained from either computed tomography (CT) or magnetic resonance imaging (MRI). During the last 10 years, many attempts have been made to optimize and automate the treatment decision, but most have been either simply time-based or applying simple population wide thresholds2. This project aimed to provide automatic decision support in a personalized manner, utilizing a deep neural network with high representational power.Methods:
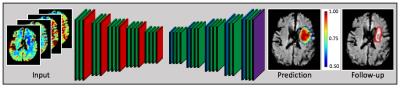
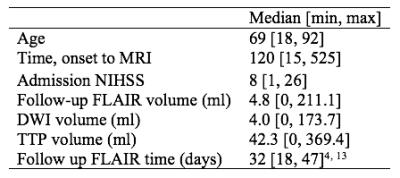
234 acute ischemic stroke patients from the I-KNOW multicenter3 (117) and perconditioning4 (117) studies were randomly divided into training (198) and test (36) sets. Figure 1 shows patients characteristics. The acute MRI protocol included diffusion-weighted imaging (DWI), T2 fluid-attenuated inversion recovery (T2-FLAIR) imaging, and gradient-echo (GE) dynamic susceptibility contrast perfusion-weighted imaging with echo time 30-50 ms. and repetition time 1500 ms. Gadolinium contrast dose 0.1 mmol/kg at injection rate 5mL/second was followed by a saline chaser. Further details are listed in Hansen et al5. Stroke evolution is physiologically highly complex and therefore an approach based on novel advances in so-called deep learning was taken. The deep learning method Convolutional Neural Network6 (CNN) have shown promising results in encoding complex imaging information at increasing levels of abstraction. CNNs consist of a number of layers of operational nodes connected in a hierarchy. The layers possess different properties and are designed together with the network architecture.
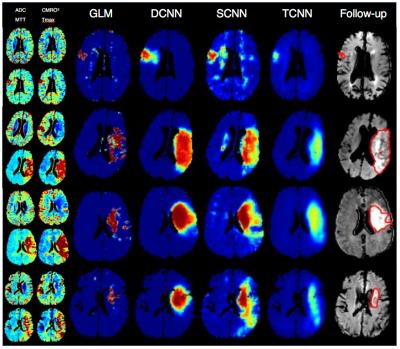
We designed a deep CNN7 (DCNN) consisting of 37 layers. To investigate performance of the deep architecture we also fitted a shallow network (SCNN) with three layers. A more advanced version of the traditional Tmax population wide thresholding method, a CNN with four layers and a linear classifier (TCNN)8, was also trained. The resulting predictive models were evaluated based on predictive performance as quantified by a combined AUC, calculated as the mean of the AUC inside and outside the manually drawn acute time to peak lesion9, which is a measurement of hypo-perfusion. A paired t-test was performed to compare AUCs from the models. A visual inspection examined the contrast and how similar the predictions were to the final infarct mask as drawn on the follow-up T2FLAIR scan. Predictions from DCNN and SCNN were based on the perfusion metrics: mean transit time (MTT) and cerebral metabolism of oxygen10 (CMRO2), and the diffusion metric: apparent diffusion coefficient (ADC). TCNN’s predictions were based on Tmax. See Figure 1 for an architectural overview of DCNN and Figure 2 for a comparison of the predictive results, which also includes the current state-of-the-art Generalized Linear Model method11, 12.
Results and discussion:
The paired t-test showed a significant difference between DCNN and GLM ($$$p<0.0001$$$), DCNN and TCNN ($$$p=0.0015$$$), and DCNN and SCNN ($$$p=0.0185$$$). Significant differences were also shown between SCNN and both GLM and TCNN ($$$p<0.0001$$$ and $$$p=0.0256$$$). The comparison of predictive models (Figure 3) showed a clear advantage of using a deep CNN to produce predictions with more layers in the network yielding a higher AUC and a better contrast. The results clearly indicate that the stacking of simple structures to deep CNNs catalyzes the model’s learning ability, whereas shallow architectures fail to capture the underlying stroke dynamics. Even a shallow CNN has the advantages of being able to retain non-linear information, making its predictions more accurate compared to a logistic regression based method such as GLM, which only retains linear information. Our DCNN was showed to be superior to the Tmax based approach and hence superior to simple Tmax thresholding.Conclusion and future work:
The new model paradigms have been shown to lead to improved predictions and thereby a much-increased potential to be used in an automated decision support system providing recommendations for personalized treatment and hopefully better outcome for the individual patient. The CNN’s will likely benefit from increasingly larger image collections, in contrast to less deep architectures, such as GLM and support vector machines, which lack the information encoding capabilities of CNNs, thereby continuing to learn with every new patient.
Acknowledgements
No acknowledgement found.References
1. Saver JL. Time is brain - quantified. Stroke. 2006;37:263-266
2. Austein F, Riedel C, Kerby T, Meyne J, Binder A, Lindner T, et al. Comparison of perfusion ct software to predict the final infarct volume after thrombectomy. Stroke. 2016;47:2311-2317
3. I-KNOW. Integrating information from molecules to man: Knowledge discovery accelerates drug development and personalized treatment in acute stroke. 2006
4. Hougaard KD, Hjort N, Zeidler D, Sorensen L, Norgaard A, Thomsen RB, et al. Remote ischemic perconditioning in thrombolysed stroke patients: Randomized study of activating endogenous neuroprotection - design and mri measurements. Int J Stroke. 2013;8:141-146
5. Hansen MB, Nagenthiraja K, Ribe LR, Dupont KH, Ostergaard L, Mouridsen K. Automated estimation of salvageable tissue: Comparison with expert readers. J Magn Reson Imaging. 2016;43:220-228
6. LeCun Y, Boser B, Denker JS, Howard RE, Habbard W, Jackel LD, et al. Handwritten digit recognition with a back-propagation network. Advances in neural information processing systems 2. 1990:396-404\
7. Badrinarayanan V, Kendall A, Cipolla R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. ArXiv e-prints. 2015;1511
8. Stier N, Vincent N, Liebeskind D, Scalzo F. Deep learning of tissue fate features in acute ischemic stroke. Ieee Int C Bioinform. 2015:1316-1321
9. Jonsdottir KY, Ostergaard L, Mouridsen K. Predicting tissue outcome from acute stroke magnetic resonance imaging improving model performance by optimal sampling of training data. Stroke. 2009;40:3006-3011
10. Jespersen SN, Ostergaard L. The roles of cerebral blood flow, capillary transit time heterogeneity, and oxygen tension in brain oxygenation and metabolism. J Cereb Blood Flow Metab. 2012;32:264-277
11. Wu O, Koroshetz WJ, Østergaard L, Buonanno FS, Copen WA, Gonzalez RG, et al. Predicting tissue outcome in acute human cerebral ischemia using combined diffusion- and perfusion-weighted mr imaging. Stroke. 2001;32:933-942
12. Wu O, Sumii T, Asahi M, Sasamata M, Ostergaard L, Rosen BR, et al. Infarct prediction and treatment assessment with mri-based algorithms in experimental stroke models. J Cerebr Blood F Met. 2007;27:196-204
13. Carrera E, Jones PS, Alawneh JA, Klaerke Mikkelsen I, Cho TH, Siemonsen S, et al. Predicting infarction within the diffusion-weighted imaging lesion: Does the mean transit time have added value? Stroke. 2011;42:1602-1607
Figures