Ashish Gupta1, Sudeep Kumar2, Shiridhar kashyap2, Deepak Kumar3, and Aditya Kapoor2
1metabolomics, Centre of Biomedical Research, Lucknow, India, 2Cardiology, SGPGIMS, Lucknow, India, 3Metabolomics, Centre of Biomedical Research, Lucknow, India
Synopsis
To address the shortcomings of the conventional biochemical approach for
the precise identification of myocardial infarction (MI), and differentiation from chronic stable angina
(CSA), and normal coronary (NC) subjects, we applied filtered serum
based metabolomics using 1H NMR spectroscopy. The study comprises
filtered sera from CAD [CSA (n=88, MI (n=90)], and NC (n=55) subjects. NMR-measured metabolites and clinical evaluation
data were examined separately using chemometric approach to probe the signature
descriptors for each cohort. This approach reveals that filtered serum based
metabolic profiling can differentiate not only NC from CSA and MI but also CSA
from MI.
INTRODUCTION
Coronary artery disease (CAD) is a global health issue. Registration of chronic stable angina (CSA) and myocardial infarction (MI) is complicated. CAD is currently diagnosed through typical angina pectoris, and with or without electrocardiogram (ECG) ST-segment and T-wave changes. Moreover, diagnosis of CSA depends solely on symptoms, which may not be classical in every patient. MI could be diagnosed with or without ECG ST-segment and T-wave changes, elevated troponin T or I levels, and expression of CK-MB. These approaches have inherent limitations (1-2). Reports suggest that metabolic biomarkers may be strongly related to outcomes and morbidity in CAD episodes (3-4). However, whether the filtered serum metabolic biomarkers may be used for CSA and MI screening is still unknown. Hence, the following potential research queries are still waiting to be observed: (1) whether NMR-derived filtered serum metabolomics are robust enough to identify subtle changes in circulating metabolites in CSA and MI. (2) whether metabolic profiling of filtered serum is able to characterize patients with CSA and MI and thus potentially join our investigative armamentarium.METHODS
All subjects underwent coronary angiography after clinical examination and biochemical evaluation for cardiac biomarkers. Conventional hs-Cardiac troponin I (TI) (hs-CTn-I), lactate dehydrogenase (LDH), and creatine kinase (CK, CK-MB, CK-MM) were measured in all subjects. Serum creatinine (SC), lipid profile [total cholesterol (TC), triglycerides (TG), very low density lipids (VLDL), high density lipids (HDL), and low density levels (LDL)] were also measured. A total of 233 subjects were enrolled in the study comprising normal coronaries (NC, n=55), CSA (n=88) and MI (n=90). Blood samples were collected and serum was separated using standard protocol. Each serum samples were passed through 3kDa filters to remove proteins and lipoproteins, and NMR experiments were performed on collected filtrates. A Bruker Avance III 800 MHz spectrometer was used to execute NMR experiments. For all the specimens, 1D 1H NMR experiments were performed by suppression of water resonance by pre-saturation. Multivariate chemometric analysis was applied on the NMR data with the help of ‘Unscrambler X’ software. The data were subjected for principal component analysis (PCA) followed by orthogonal partial least square discriminant analysis (OPLS-DA). To avoid over-fitting of the OPLS-DA model, an internal cross-validation (ICV) was also applied using 60% of the data as training set and 40% of the data as test set. A prediction model was constructed to detect potential biomarkers related to the discrimination between CAD and NC following CSA and MI cohorts. To evaluate the clinical utility of biomarkers, ROC analysis was also performed. RESULTS
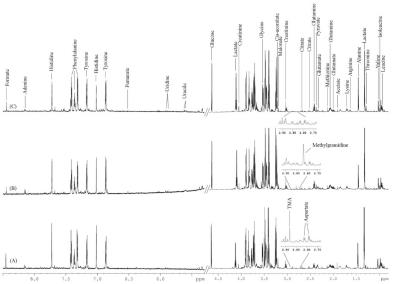
Figure 1 shows 1D 1H NMR spectra of filtered serum of NC, CSA, and MI with an overview of complete metabolic profile and chemical shift assignments. PCA and OPLS-DA screening outcomes revealed that the pattern of methylguanidine (MG), lactate, creatinine, threonine, aspartate and TMA, and TI, LDH, CK, and SC were perturbed in CAD compared to NC. Statistical analysis demonstrated high precision (93.6% by NMR and 67.4% by clinical measures) to distinguish CAD from NC. Further analysis indicated that methylguanidine, arginine, and threonine, and TI, LDH, and SC were significantly perturbed in CSA compared to MI. Statistical analysis demonstrated high accuracy (88.2% by NMR and 92.1% by clinical measures) to discriminate CSA from MI.DISCUSSION
The main finding
of this study is that (i) NMR-derived serum metabolic
phenotypes are sufficiently robust to detect
subtle changes in circulating metabolites in CSA and MI; (ii) metabolomic
outcomes are comparable to clinical measures and are rapid, robust, and
unambiguous; thus, metabolomics approach has the potential to join our investigative armamentarium. In CSA, the
ischemia-reperfusion phenomenon causes the release of free radicals and
pro-inflammatory mediators (5) which stimulate neutrophils for MG
production from creatinine, causing oxidative stress (5-7). Augmented level
of threonine in MI is to reduce necrotic injury-causing myocardial
ischemia-reperfusion and the generation of oxidative stress (8). Augmented
level of arginine in CSA and MI establishes the role of oxidative stress in CAD (6).
Under hypoxia or ischemia pathology, an augmented
reliance on anaerobic myocardial metabolism takes place that escalates the
release of lactate (9). Reduction in aspartate and histidine level
in CAD; concurs with a previous observation of acute coronary syndrome (10-11). Altered level of TMA in CAD concurs that TMA is mechanistically related
to atherosclerosis and associated risk of cardiovascular disease (12). In essence, our finding reveals that 1H NMR based metabolomics of filtered serum appears to
be a robust, rapid, and minimally-invasive approach to probe CSA and MI.
Acknowledgements
No acknowledgement found.References
(1) Circulation
1999; 99: 1671-1677. (2) Clin Chim Acta 1997; 257:
99-115. (3) Nature Rev Drug Disc 2003; 2: 668-676. (4) Nature 2004; 429: 188-193. (5) J Clin
Invest 2005; 115: 500-508. (6) Clin Exp Nephrol 2000;
4: 231-235. (7) Nephron 1994; 66:
140-146. (8) Annu Rev Physiol 2010;
72: 19-44. (9) J Thorac Cardiovasc Surg 1976;
71: 726-735. (10) Anal Bioanal Chem 2009; 394: 1571-1524. (11) Circ Cardiovasc
Genet 2015; 8: 351-355. (12) J Clin Invest 2014; 124: 4204-4211