5532
Spatial Hadamard encoding of J-edited spectroscopy using slice-selective editing pulses1Biomedical Engineering, Johns Hopkins School of Medicine, Baltimore, MD, United States, 2F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins School of Medicine, Baltimore, MD, United States
Synopsis
A new approach for simultaneous dual-voxel J-difference spectral editing is described, that uses spatially selective spectral-editing pulses and Hadamard encoding. The theoretical framework for Spatial Hadamard Editing and Reconstruction for Parallel Acquisition (SHERPA) was developed, applying gradient pulses during the frequency selective editing pulses. SHERPA was simulated for GABA, tested in a two-compartment GABA phantom, and applied to the left and right hemispheres of ten normal subjects. SHERPA was successfully implemented with results in close agreement with conventional MEGA-PRESS scans. Compared to conventional single-voxel single-metabolite J-difference editing, two-fold acceleration is possible without significant loss of SNR using the SHERPA method.
Purpose
It is often desirable to measure spectra from multiple brain regions, but sequential collection of single-voxel MRS data is time-prohibitive, and inefficient. A new approach for simultaneous multi-voxel detection using J-difference editing is described. The method, ‘Spatial Hadamard Editing and Reconstruction for Parallel Acquisition’ (SHERPA), uses multi-slice RF pulses, and a field gradient during the editing pulses to encode location-dependent editing. The editing pulses are applied in a spatial Hadamard editing scheme, allowing difference-edited spectra from two or more voxels to be reconstructed separately.Methods
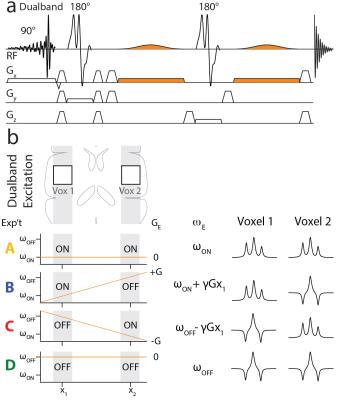
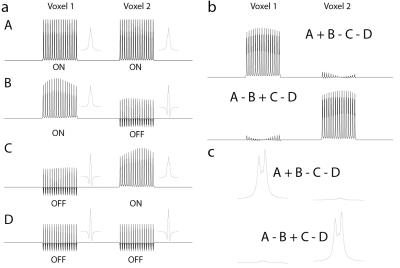
Starting from the MEGA-PRESS (1) sequence, SHERPA uses a dual-band excitation pulse to excite two (or more) voxels simultaneously and adds a field gradient during the editing pulses (Figure 1a), allowing position-dependent control of the editing pulse frequencies (Figure 1b). For 2 voxels, 4 experiments are performed with the editing pulses either on-resonance (ON) or off-resonance (OFF) to the editing target as demonstrated for GABA in Figure 1: A (ONvox1, ONvox2); B (ONvox1, OFFvox2); C (OFFvox1, ONvox2); and D (OFFvox1, OFFvox2) (Figure 1b). Experiments A and D require the same editing frequency in both voxels, and thus have no editing gradient applied. Experiments B and C require different editing pulse frequencies in the two voxels, and have editing gradients with opposite polarity. Combining the experiments, A+B-C-D and A-B+C-D give the J-difference-edited spectrum for Voxel 1 and Voxel 2 respectively. All experiments were performed on a Philips Achieva 3T scanner. Spatially-resolved simulations in the excitation direction were performed using ‘FID-A’ (2). GABA-edited SHERPA was simulated for two voxels 30 mm wide and 66 mm apart, TE=80 ms, and editing frequencies: ON 1.9 ppm and OFF 1.0 ppm. SHERPA experiments were performed in a 1L agarose gel phantom (pH 7.2) with different GABA concentrations in the top (10 mM) and bottom (16 mM) halves, with TR 2 s; 2.7x2.7x2.7 cm3 voxels; and 64 signal averages. SHERPA experiments were acquired with both voxels excited and with only Voxel 1 or Voxel 2 excited. MEGA-PRESS acquisitions for each voxel were also collected. To acquire non-water-suppressed data for each voxel for quantification, SHERPA was performed with editing pulses applied to the water resonance (ON 4.68 ppm, OFF 5.25 ppm) which allows for the separation of water signals from each voxel. SHERPA GABA editing was performed in 10 healthy adults (3 female, 7 male; age 25 ± 3 years) in 3x3x3 cm3 voxels in the left and right hemispheres 66 mm apart, with 320 averages acquired and 2nd order shimming. GABA MEGA-PRESS acquisitions of each voxel separately were also acquired in one subject (1 male, age 28) for comparison. In-house software was used to combine the signals from the different channels and the GABA concentrations were estimated using the ‘Gannet’ program (3).Results
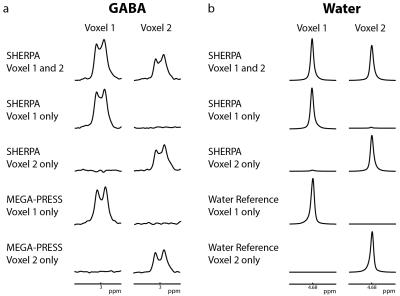
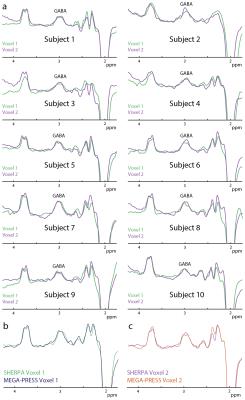
Simulated GABA sub-spectra (A,B,C,D) as a function of position are shown in Figure 2a and show the expected modulation pattern. Variation of the ON signal across the voxel is fairly small in B and C. Hadamard combinations of the sub-experiments (Figure 2b, 2c) demonstrate good separation of the GABA signal from each voxel (2.2% crosstalk). Figure 3a shows GABA-edited SHERPA phantom spectra. There is good agreement between the SHERPA and MEGA-PRESS spectra with minimal crosstalk. The GABA concentration ratio (voxel 1/voxel 2) was 1.61:1 for SHERPA, 1.68:1 for single-voxel SHERPA, and 1.67:1 for MEGA-PRESS, and all are close to the actual concentration ratio of 1.6:1. Figure 3b shows the reconstructed water spectra from corresponding SHERPA and MEGA-PRESS acquisitions are consistent. Figure 4a shows that the GABA-edited spectra are comparable between voxels in the left and right hemispheres, and between subjects. GABA concentrations estimated by water referencing were: 3.25 ± 0.44 (institutional units, i.u.) for voxel 1 and 3.05 ± 0.54 i.u. for voxel 2. Figure 4b and 4c show that the in vivo GABA-edited SHERPA spectra are comparable to those from the standard MEGA-PRESS acquisition.Discussion
SHERPA is capable of editing two different brain regions simultaneously using spatially dependent spectral editing pulses, which provides a two-fold acceleration over sequential MEGA-PRESS measurements. There is little crosstalk between the two voxels and minimal loss of signal compared to conventional MEGA-PRESS scans. SHERPA should also be applicable to any other J-coupled molecules that can be edited with MEGA-PRESS, and can be applied to other combinations of brain regions.Acknowledgements
This work was supported by NIH R01 EB016089 and P41 EB015909.References
1.
Mescher M, Merkle H, Kirsch J, Garwood M,
Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR
Biomed 1998;11:266–272.
2.
Simpson R, Devenyi GA, Jezzard P, Hennessy TJ,
Near J. Advanced processing and simulation of MRS data using the FID appliance
(FID-A)-An open source, MATLAB-based toolkit. Magn Reson Med 2015.
3.
Edden RAE, Puts NAJ, Harris AD, Barker PB, Evans
CJ. Gannet: A batch-processing tool for the quantitative analysis of
gamma-aminobutyric acid-edited MR spectroscopy spectra. J Magn Reson Imaging
2013;40:1445–1452.
Figures