5531
High resolution cortical spectroscopy at 7T using lipid signal crushing and a high density receive array.1Radiology, University Medical Center Utrecht, Utrecht, Netherlands, 2Psychiatry, University Medical Center Utrecht, Utrecht, Netherlands
Synopsis
In this work we attempt to overcome MRSI limitations associated with extra-cranial lipid signal leakage and low SNR at high resolution. We use a dedicated crusher coil for lipid signal removal, in combination with a high density receive array and an 7T MR scanner for boosted SNR.
Introduction
Neurodegenerative diseases and mental disorders are thought to cause changes in metabolic processes related to neuronal functioning in the cerebral cortex. Elevated glutamate-glutamine levels have been reported in un-medicated schizophrenia patients(1), while changes in metabolite levels of N-acetyl aspartate, creatine and myo-inositol have been associated with Alzheimer’s related neurodegeneration(2). Cortical metabolite concentrations can be detected using proton spectroscopy (H1-MRS), however, limitations relating to metabolite signal contamination from extracranial lipids and partial volume effects relating to the large voxel sizes have hindered the progress of magnetic resonance spectroscopic imaging (MRSI) as a routine clinical application. In this work we attempt to overcome these limitations through the use of a crusher coil for lipid signal removal, in combination with a high density receive array and an 7T MR scanner for boosted SNR.Methods
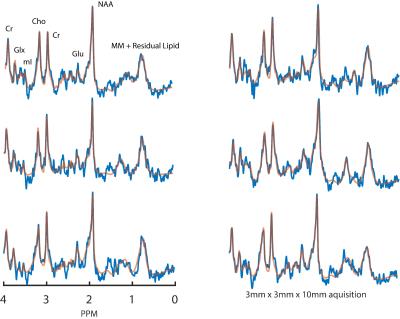
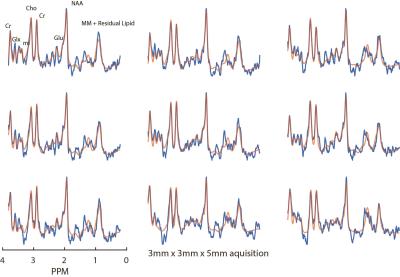
Two datasets were acquired from a healthy 35 year old male subject with voxel dimensions of 3x3x10mm3 and 3x3x5mm3, respectively. MRSI data were acquired using a Philips 7T scanner (Best, The Netherlands) and dual channel volume transmit coil (NOVA medical, Wilmington MA, USA) in combination with a high density, 32 channel receive array(3) (MR Coils, Zaltbommel, NL). Extracranial lipid signal contamination was reduced using a Crusher Coil(4) (MR Coils) which was pulsed directly after each RF excitation for approximately 0.8ms; for setup see fig. 1. A pulse acquire sequence was used with a CHESS water suppression. For scan parameters see figure legends. An un-suppressed water acquisition (at half the spatial resolution) was used for eddy-current correction and for removal of residual water signal and water side bands. The water spectrum was fitted to the water suppressed spectrum and the fit was subtracted and followed by SVD filtering. The resulting spectra were phased and fitted using LCModel (5).Results
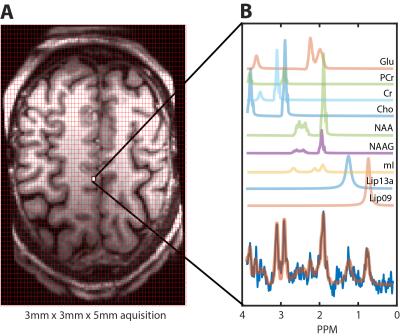
Example, single voxel spectra derived from the lower resolution acquisition are shown in figure 2. Average SNR and FWHM for this selection were 6.83±0.98 and 12.75±1.73Hz, respectively. Figure 3 shows a 3x3 grid of adjacent voxels selected from a predominantly GM region atop the cortex. Average SNR and FWHM for this selection were 3.56±0.53 and 16.89±2.58Hz, respectively. These results are in line with expectations since halving voxel volume should yield a comparable reduction in signal. Results from fitting metabolite peaks to an example spectra using LCModel, as well as their representative GM location are show figure 4.Discussion
Using a crusher coil to suppress extra-cranial lipid signal and a high density receive array, we are able to acquire high resolution MRSI datasets of cortical tissue. Our work raises the potential for high resolution spectroscopy of the cortex at 7T; previous studies have reported voxel dimensions of 3x3x10mm3 (4) and 3.4x3.4x8mm3 (6). Importantly, we are able to suppress high lipid signal during the aquisition and acquire data within a reasonable scan time on the order of 12 minutes. Apart from reduced partial volume effects, high resolution MRSI has intrinsically reduced lipid contamination. The spatial extent of the point spread function side lobes for each voxel is lower. In combination with external lipid signal crushing, cortical metabolite spectra are therefore less susceptible to corruption from signal leakages originating at extra-cranial lipid regions. Several issues require further investigation; particularly the optimal current amplitude and duration for lipid signal removal considering the limited penetration depth of the surface receive coils.Conclusion
Initial results are promising and we have shown that it is possible to acquire good quality in voxels smaller than customary for MRSI imaging. Refinement of this methodology brings the potential to further increase MRSI resolution to gain insight into the mechanisms of cortical metabolic disorders and fine scale neuro-chemical responses.Acknowledgements
No acknowledgement found.References
1. Poels EMP, et al., Imaging glutamate in schizophrenia: review of findings and implications for drug discovery. Mol Psychiatry 2014;19(1):20-29.
2. Hancu I, et al., 1H MR spectroscopy using TE averaged PRESS: a more sensitive technique to detect neurodegeneration associated with Alzheimer's disease. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine 2005;53(4):777-782.
3. Petridou N, et al., NMR in biomedicine 2013;26(1):65-73.
4. Boer VO, et al., Lipid suppression for brain MRI and MRSI by means of a dedicated crusher coil. Magnetic resonance in medicine 2015;73(6):2062-2068.
5. Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magnetic resonance in medicine 1993;30(6):672-679.
6. Strasser B, et al., (2 + 1)D-CAIPIRINHA accelerated MR spectroscopic imaging of the brain at 7T. Magnetic resonance in medicine Aug 22. doi: 10.1002/mrm.26386
Figures