5483
Impact of time sample selection and model function design on the quantification of fatty acid composition: in vitro and in vivo studies.1Univ. Lyon, INSA‐Lyon, Université Claude Bernard Lyon 1, UJM-Saint Etienne, CNRS, Inserm, CREATIS UMR 5220, U1206, F69621, Lyon, France, 2INSERM, UMR 1099, Rennes, France, 3Univ Rennes 1, LTSI, Rennes, France, 4Department of Physiology, Faculty of Biology and Medicine, University of Lausanne, Lausanne, Switzerland, 5Centre de Recherche en Nutrition Humaine Rhône-Alpes (CRNH-RA), Centre Hospitalier Lyon Sud, Pierre-Bénite, Lyon, France, 6Hospices Civils de Lyon, Département d'imagerie digestive, CHU Edouard Herriot, Lyon, France
Synopsis
Interest in the follow-up of fatty acid composition(saturated, polyunsaturated and monounsaturated fatty acid) in the body is growing. Quantitative MR spectroscopy can give access to this fat composition. Today, several quantification methods are used (e.g LCModel, AMARES). However the statistical outcome issued from a quantitative analysis of the lipid signal can be greatly influenced by the used quantification method. We analyze 1) the impact of the time sample selection and design of the model function on the parameter identifiability 2) the quantification results obtained with different quantification models on acquisitions performed in vitro (oils) and in vivo (subcutaneous adipose tissue).
Purpose
Interest in the fat quantification has grown in recent years. In particular, the nutrition studies are interested in the variation of fatty acid composition in the body: saturated fatty acid (SFA), polyunsaturated (PUFA) and monounsaturated (MUFA). Magnetic resonance spectroscopy (MRS) is a quantitative technique that gives access to this fat composition. At this time, several quantification methods, primarily developed for brain MRS analysis, are used (e.g LCModel, AMARES). However, it has been recently described that, in the case of lipid signal, the statistical outcome issued from a quantitative analysis can be greatly influenced by the used quantification method1.
In the present work, we investigate the particular case of lipid quantification in adipose tissue; for which the lipid components are predominant and we study the impact of the time sample selection and design of the model function on the parameter identifiability (given by the condition number of the Jacobian Matrix2) and on the parameter relative Cramer Rao Lower Bound (CRLB). These two criteria give insight about the ability of the parameter estimation to provide precise inference. We then analyze the quantification results obtained with different quantification models- adding results from the reference LCModel method- on in vitro oil and in vivo subcutaneous adipose tissue (AT) acquisitions.
Materials and methods
In vitro MRS was performed on a preclinical 4.7T BioSpec Bruker system, a PRESS sequence was used with TR = 5000 ms, TE = 7.89 ms, VOI of 4×4×4 mm3, 32 signal averages; on 10 commercial oils.
In vivo MRS was carried out on Philips Ingenia 3T system the STEAM sequence was used with TR= 3000ms, TE= 14ms, VOI of 20×20×20 mm3, 4 signal averages and 1024 data points. On five subjects, an MR spectrum was acquired twice (for test retest) in the abdominal subcutaneous AT using respiratory triggering.
Most of the MRS quantification methods are based on a model function to be adjusted to the acquired data. When fitting in the time domain the time sample used in the fitting procedure can be selected.
Studied Model functions.
The first studied model function is inspired from a quantification method applied on multi-gradient echo imaging3 (Eq.1). $$Eq.1:f(t)=\left(A_w*n_{water}+A_f*\sum_{k=1}^8n_k(ndb,nmidb)*e^{2i{\pi}f_kt}\right)e^{-\frac{t}{T2^*}}$$ The second model is based on a Voigt Model4 (Eq.2) close to AMARES signal description.$$Eq.2:f(t)=e^{i\phi_0}*\sum_{k=1}^9c_ke^{\alpha_kt+\left(\beta_kt\right)^2+2i{\pi}f_kt}$$
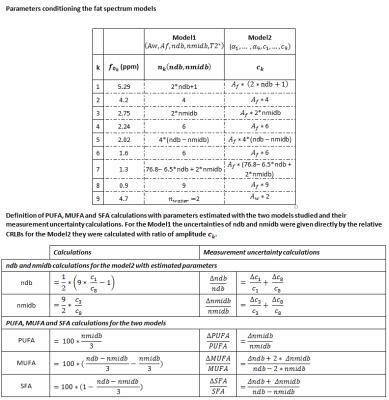
In the case of the lipids, the relative amplitudes of the peaks are linked (see Table1). These relations can be introduced in the model (Model1) or deduced from the fitted component amplitude (Model2).
Undersampling of
temporal signal within the fitting procedure was
tested
with $$$t=n*t_e$$$ with $$$t_e=\frac{1}{2*(4.7-1.3)*B_0*\frac{\gamma}{2\pi}}$$$, the results presented below were for n=32.
Theoretical model comparison
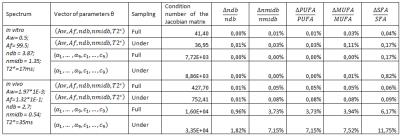
For each model function and each case (oils and in vivo), some realistic parameter values were used and the condition number of the Jacobian matrix (cond-J), and the parameter uncertainty, $$$\frac{\triangle\theta}{\theta}$$$, as the relative the CRLBs, were computed ($$$\frac{\triangle\theta}{\theta}=\sigma_0*\frac{\sqrt{F(\theta)^{-1}}}{\theta}$$$ where $$$\theta=\left\{ndb,nmidb,c_1,c_3,c_8\right\}$$$, $$$F^{-1}=\left[J^T.J\right]^{-1}$$$ the inverse Fisher matrix and $$$\sigma_0$$$ standard deviation of noise, for PUFA, MUFA and SFA the calculation is detailed in table1.
Five estimated parameters $$$(A_w,A_f,ndb,nmidb,T2^*)$$$ for Model1 and 2*9 estimated parameters $$$(\alpha_1,...,\alpha_9,c_1,...,c_9)$$$ for Model2 were considered. Indeed, to be sure that the two models describe mathematically the same signal we used for Model2: $$${\phi_0}=0$$$, $$${\alpha_k}=-\frac{1}{T2^*}$$$, $$$ {\beta_k}=0$$$, $$$c_k=A_f*n_k(ndb,nmidb)$$$ for $$$k=\left\{1,...,8\right\}$$$, $$$c_9=A_w*n_{water}$$$ and $$$f_k=\left(f_{0_k}-4.7\right)*B_0*\frac{\gamma}{2\pi}$$$.
Quantification results comparison
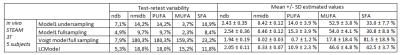
In vivo and in vitro MR spectra were quantified, according to a least-square fit implemented in a home-made MATLAB software, with the Model1 and Model2 (full sampling and undersampling) as well as with LCModel. For the oils, quantification results are compared to the theoretical known oil composition. For the AT, the variability percentage of the test-retest for each index was calculated.
Results
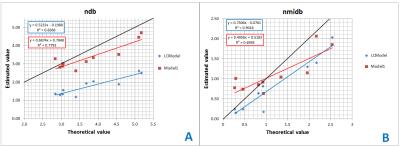
Results on the theoretical model comparison are summarized in Table2. For oil quantification results, LCModel showed the best correlation with theoretical values but under-estimated ndb. Model1 showed also good correlation and estimated values in agreement with the theoretical values when employing the undersampling (Figure1). For the in vivo results (Table3), the test-retest variability was the smallest with Model1 fullsampling. LCmodel shows good test-retest variability but with different estimated parameter values. Figure2 shows the same spectrum fitted with two different models who lead to different results despite both showing small residual.Discussion/conclusion
This work demonstrates that, in the particular case of the lipid MRS quantification, the model function simplification (here by linking, from prior knowledge, the peak amplitude) appear to be key leverage point to increase the precision of the result. Constraints and prior knowledge as well as the time sampling selection also seem to impact the bias of the estimation and need to be further investigated.Acknowledgements
This work was performed within the framework of the LABEX PRIMES (ANR-11-LABX-0063) of Université de Lyon, within the program "Investissements d'Avenir" (ANR-11-IDEX-0007) operated by the French National Research Agency (ANR), IHU Opera et PHRC-IR Visfatir.References
1Mosconi E, Sima DM, Osorio Garcia MI, Fontanella M, Fiorini S, Van Huffel S and Marzola P. Different quantification algorithms may lead to different results: a comparison using proton MRS lipid signals. NMR Biomed (2014). 27:431-443.
2Reid J G, Structural identifiability in linear time-invariant systems. IEEE Trans. Automat. Control. (1977). 22:242–246.
3Leporq B, Lambert SA, Ronot M, Vilgrain V, Van Beers BE. Quantification of the triglyceride fatty acid composition with 3.0 T MRI. NMR Biomed (2014). 27(10):1211-21.
4Ratiney, H., A. Bucur, M. Sdika, O. Beuf, F. Pilleul, and S. Cavassila. Effective Voigt Model Estimation Using Multiple Random Starting Values and Parameter Bounds Settings for in Vivo Hepatic 1H Magnetic Resonance Spectroscopic Data. In 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro (2008). ISBI 2008, 1529–32, 2008.
5Hamilton G, Yokoo T, Bydder M, Cruite I, Schroeder ME, Sirlin CB, Middleton MS. In vivo characterization of the liver fat 1H MR spectrum. NMR Biomed (2011). 24: 784–790.
6Garaulet M, Hernandez-Morante JJ, Lujan J, Tebar FJ, Zamora S. Relationship between fat cell size and number and fatty acid composition in adipose tissue from different fat depots in overweight/obese humans (2006).International Journal of Obesity. 30(6):899-905.
Figures