5345
Inflated false positive rates in fMRI depend on the voxel size of normalized images1Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 2Department of Biomedical Magnetic Resonance, University Hospital Tübingen, Tübingen, Germany, 3Magnetic Resonance Center, Max Planck Institute for Biological Cybernetics, Tübingen, Germany
Synopsis
Recently, Eklund et al published a manuscript discussing the issue of false positive results in functional MRI (fMRI) using the most common software packages. In their analysis, image upscaling was performed in fMRI preprocessing after registering images into a standard space (normalization). We show that the degree of image upscaling used for normalization impacts the statistical results when using the Gaussian Random field approach. A higher upscaling generally leads to smaller p-values increasing the number of false positive clusters. This result is quite troubling because statistical inference should not depend on a preprocessing parameter which can be chosen ad libitum.
Introduction
In a recent manuscript, Eklund et al.1 reported inflated false positive rates in functional magnetic resonance imaging (fMRI) using common software packages including SPM, FSL, and AFNI. Briefly, a nominal family-wise error rate of 5% in the parametric statistical evaluation was shown to be conservative for voxel-wise inference but not for cluster-wise inference. As a cause of the observed invalid cluster inferences, the authors suggested that the spatial autocorrelation functions do not follow the assumed Gaussian shape.
Here, we would like to draw attention to an important aspect that was not addressed in this publication. Specifically, we note that statistical inferences obtained using the Gauss random field approach depend heavily on a pre-processing parameter that was not included in the analysis performed by Eklund et al.1, namely the spatial resolution to which the data are resampled and interpolated during pre-processsing. This resampling is needed to align the data to a common anatomical template. Eklund et al.1 used the common default setting of 2×2×2 mm³. In response to the paper by Eklund et al., Flandin and Friston2 used a different setting of this parameter, namely 3×3×3 mm³. Together with a more stringent initial cluster-forming threshold, they did not observe inflated false positive rates. However, a spatial resolution of 2×2×2 mm³ is the default value in two major software packages (SPM, FSL) and, hence, it is likely to be used for processing fMRI data by these packages. Moreover, in previous work, Friston and colleagues3 stated that resampling to 2×2×2 mm³ renders the analysis “more sensitive”. It is, thus, unclear what a valid setting for this parameter should be. Therefore, it is of substantial relevance to systematically assess its influence on statistical inference.
Methods
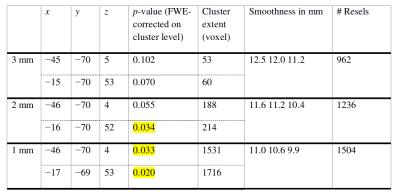
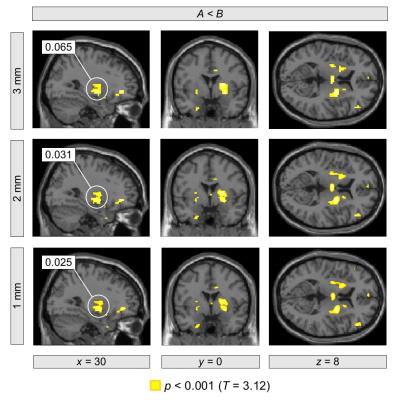
We analyzed 47 resting-state fMRI data sets, each acquired at a nominal spatial resolution of 3×3×4 mm3 with 300 volumes. Using a strategy analogous to that of Eklund et al. we imposed various fake designs including block- and event-related types. We tested the following resampling parameters: 3×3×3 mm3, 2×2×2 mm3, and 1×1×1 mm3. Using SPM12 with family-wise error (FWE) correction for multiple comparisons based on the Gauss random field approach, we first evaluated each data set separately. Table 1 and Figure 1 illustrate a typical result. With higher resampling resolutions, we found that the FWE-corrected p-values decreased systematically leading to a concomitant increase in false positives. Similar systematic effects were obtained in most of the 47 data sets. Furthermore, we performed a group-level inference with pooling of all 47 data sets. Again, we observed that the FWE-corrected p-values decreased systematically with higher resampling resolutions.Conclusion
It appears that there is a systematic dependence of the false positive rate on the resampling parameter with smaller voxel sizes leading to smaller FWE-corrected p-values and, thereby, more false positives. While some dependence on pre-processing parameters may be inevitable, a systematic dependence of this type is clearly worrisome, because researchers may be tempted to interpolate their data until the desired statistical significance level is reached. Statistical inference should certainly not depend in such a systematic way on a pre-processing parameter that can be set ad libitum. Clearly, this issue requires further in-depth analysis.Acknowledgements
No acknowledgement found.References
1. Eklund A, Nichols TE, Knutsson H. Cluster failure: Why fMRI inferences for spatial extent have inflated false-positive rates, Proc Natl Acad Sci U S A. 2016;113(28):7900-7905. doi:10.1073/pnas.1602413113
2. Flandin G, Friston KJ. Analysis of family-wise error rates in statistical parametric mapping using random field theory, arXiv 2016;1606.08199v1 [stat.AP], 27 Jun 2016, http://arxiv.org/pdf/1606.08199.pdf
3. Hopfinger JB, Büchel C, Holmes AP, Friston KJ. A study of analysis parameters that influence the sensitivity of event-related fMRI analyses, Neuroimage 2000;11(4):326-333. doi:10.1006/nimg.2000.0549
Figures