5336
A New Method of HRF Estimation Containing High Frequency Content1Cleveland Clinic Lou Ruvo Center for Brain Health, Las Vegas, NV, United States, 2University of Colorado Boulder, Boulder, CO, United States
Synopsis
A new HRF estimation method was introduced to improve the accuracy in recovering HRFs with wider frequency range. This non-smooth optimization problem was solved via BFGS technique. Results from simulated data demonstrate the accuracy and reliability of this new method in recovering HRF. Results from an event-related fMRI dataset further demonstrate the ability of the proposed method in capturing the variation of HRFs across different brain regions and subject populations.
Introduction
In
functional magnetic resonance imaging (fMRI), the measured signal is assumed to
be a linear convolution of neuronal response and the hemodynamic response
function (HRF)1. The variability of HRF has been widely recognized2
and various models of the evoked HRFs have been investigated3,4.
However, current models involve simplified estimations of HRF and focus on
limited frequency ranges, therefore, they are not suitable to analyze fMRI data
with a higher frequency band. In this study, we proposed a new HRF estimation
method, where we set a larger range of HRF parameters to cover wider frequency range
and explicitly include a regularization term to penalize the curvature of the
HRF and guarantee a smoothed estimated function. The new method is validated
with both simulated data and real event-related fMRI data.Methods
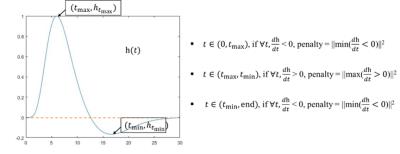
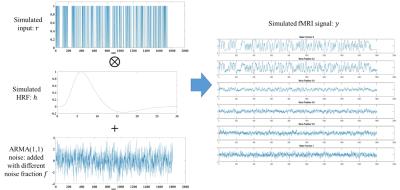
Model: We parameterized the estimated HRF model as a combination of the canonical HRF and its first order temporal derivative:$$$h(t) = (t^{a_2}-a_3t^{a_4})+a_5(e^{-t}(a_2t^{(a_2-1)}-a_3a_4t^{(a_4-1)})-e^{-t}(t^{a_2}-a_3t^{a_4}))$$$, where $$$a_2,a_3,a_4,a_5$$$ and are the unknowns of $$$h$$$. Let $$$y$$$ be a vector representing the time course from one voxel and $$$r$$$ represents n functions used to model the fMRI task design, the optimization problem here is: $$$||y-r*h||^2+\lambda\times$$$penalty, where $$$*$$$ denotes convolution. In Fig. 1, we explained the penalty term we used in detail. This non-smooth optimization problem was solved with the Broyden-Fletcher-Goldfarb-Shanno (BFGS) algorithm and inexact line search method5. A wide range of possible HRFs were generated and then clustered into 100 clusters based on their shape profiles using k-means clustering. The corresponding parameters of each cluster center were used as 100 starting points of BFGS algorithm. Simulation: A sequence of inputs with same amplitude and a duration of 3 seconds were generated as hypothetical task events (Fig. 2, left top). These events were then convolved with a hypothetical HRF (Fig. 2, left middle). The resultant signal was downsampled to mimic a TR of 0.765 seconds (same as our real fMRI data). Autoregressive moving average model (ARMA (1, 1))6 noise (Fig. 2, left bottom) was then added to the downsampled signal with different noise fractions to obtain simulated fMRI signals at different noise levels (Fig. 2, right). Imaging: Functional MRI was performed on 5 healthy subjects (age: 18 to 22) on a 3T Trio Tim Siemens MRI scanner (32-channel head coil, multiband accelerate factor = 8, TR/TE/flip angle = 765ms/30ms/44deg, 80 slices in oblique coronal orientation perpendicular to long axis of hippocampus, resolution 1.65mm x 1.65mm x 2mm, BW = 1724 Hz/pixel, echo spacing = 0.72 ms and 2380 time frames). An event-related repetition suppression spatial memory task was performed, involving viewing pairs of visual objects presented for the first time, repetitions of the same objects, or lures that were visually similar to the first-time objects. A control task (pixelated background of the math task) occurred occasionally requiring subjects to perform simple addition. Each task was on for three seconds and the blank between tasks was one second. Activations in the hippocampus were located and HRFs were estimated for every voxel in each hippocampal subregions.Results
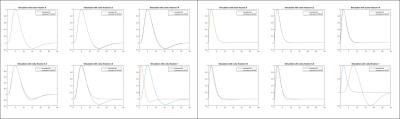
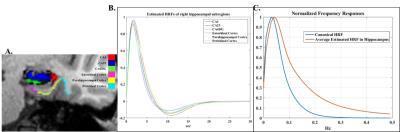
Fig. 3 shows two simulated HRFs (blue), at low and high frequency band, respectively, and the estimated results (red) obtained from simulated signals with different noise levels. For simulations with noise fraction 80%, the maximum difference between estimated HRF and simulated HRF is 16.28%. A full run of possible HRFs are tested and the highest estimation difference is less than 20% (when 80%). In real fMRI spatial memory dataset, bilateral activations in the hippocampus are observed for every contrast of memory v/s control. Hippocampal subfields and the estimated HRFs of each subregions, averaging over all active voxels are shown in Fig. 5 (A) and (B), respectively. Frequency responses of the canonical HRF and the obtained estimated HRF are shown in Fig. 5(C).Discussion
Simulation results indicate that the proposed method is able to accurately recover HRFs with wider frequency band (Fig. 4). In validation with real event-related fMRI data, estimated HRF in hippocampal subregions shows a much shorter time to peak (Fig. 5(B)) and a higher frequency band (Fig. 5(C)), as compared to the canonical HRF. This may due to the subjects recruited in this study are relatively younger.Conclusion
A new method of estimating HRF is proposed and solved with optimization techniques. Results from simulated data demonstrate the reliability of this method. Results from an event-related fMRI dataset further demonstrate the ability of the proposed method in capturing the variation of HRFs across different brain regions and subject populations.Acknowledgements
The study was supported partially by the National Institutes of Health (grant number 1R01EB014284) and by National Institute of General Medical Sciences (grant number P20GM109025).References
[1]. Ogawa S, et al. Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging. PNAS 1992, 89:5951-5955.
[2]. Handwerker D.A, et al. Variation of BOLD hemodynamic responses across subjects and brain regions and their effects on statistical analyses. Neuroimage 2014, 21(4): 1639-1651.
[3]. Buxton R.B, et al. Modeling the hemodynamic response to brain activation. Neuroimage 2005, 23: S220-S233.
[4]. Lindquist M.A, et al. Modeling the hemodynamic response function in fMRI: efficiency, bias and mis-modeling. Neuroimage 2009, 45(1): S187-S198.
[5]. Lewis A.S, et al. Non-smooth optimization via BFGS. Submitted to SIAM J. Optimiz, 2009, pp.1-35.
[6]. Purdon P.L, et al. Effect of temporal autocorrelation due to physiological noise and stimulus paradigm on voxel-level false-positive rates in fMRI. Human brain mapping, 1998, 6(4): 239-249.
Figures