5289
Individualized Functional Parcellation of Human Amygdala using a Semi-Supervised Clustering Method based on 7T Resting State fMRI Data1State Key Laboratory of Brain and Cognitive Science, Beijing MR Center for Brain Research, Institute of Biophysics, Chinese Academy of Sciences, Beijing, People's Republic of China, 2University of Chinese Academy of Sciences, Beijing, People's Republic of China, 3Department of Biomedical Engineering, School of Bioinformatics, Chongqing University of Posts and Telecommunications, Chongqing, People's Republic of China, 4Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, United States, 5Beijing Institute for Brain Disorders, Beijing, People's Republic of China
Synopsis
Functional subspecialization of human amygdala has been revealed in a variety of studies based on histological, in-vivo imaging, and meta-data. However, most of the existing studies identified functional subregions of amygdala at a group level. In this study, we investigated individualized functional neuroanatomy of amygdala based on 7T resting-state fMRI data with high spatiotemporal resolution. Our results have demonstrated that an improved semi-supervised clustering algorithm successfully parcellated individual subjects’ amygdala into 3 subregions, each of them having distinctive functional connectivity patterns. The individualized functional subregions of amygdala may better capture individual variability in functional neuroanatomy than their group level counterparts.
Introduction
Anatomical and functional heterogeneity of the amygdala has been revealed in a variety of studies in animals and in human1. In-vivo neuroimaging studies of amygdala have identified distinctive functional subregions of the amygdala at a group level. However, individualized functional neuroanatomy of amygdala subregions remained less investigated, partially due to low spatiotemporal resolution of standard neuroimaging data. To identify functional subregions of the human amygdala, unsupervised clustering algorithms have been adopted to parcellate the amygdala based on standard resting-state fMRI (rsfMRI) data. However, these methods are not equipped to make use of existing knowledge of functional neuroanatomy of amygdala. In this study, we adopted a semi-supervised clustering algorithm to parcellate the human amygdala based on 7T rsfMRI data with high spatiotemporal resolution aiming to derive individualized functional parcellation of the human amygdala. We have validated our parcellation results by investigating functional connectivity (FC) patterns of the amygdala subregions.Materials and Methods
Data acquisition
Twenty healthy subjects were enrolled in this study. The study was approved by the local IRB and written informed consents were obtained from all participants. All subjects underwent T1-weighted (T1w) anatomical imaging with MPRAGE sequence and rsfMRI imaging with GRE-EPI sequence on a 7T research system (Siemens, Erlangen, Germany) equipped with a Nova Medical 32-channel head coil. All participants also underwent T1w anatomical imaging on a MAGNETOM Prisma 3T MR scanner with a 20-channel head coil. The 3T T1w scans were used to locate amygdala using FreeSurfer.
Parameters for rsfMRI imaging at 7T were as follows: TR=3000ms, TE=22ms, FA=70degree, slices=70, FOV = 192x192mm, voxel size = 1.5mm iso, measurements=200. Parameters for T1w anatomical imaging at 7T: TR= 2200ms, TE=3.21ms, voxel size=0.7mm iso; Parameters for T1w anatomical imaging at 3T: TR=2530ms, TE=3.37ms, voxel size=1.0mm iso.
Data processing
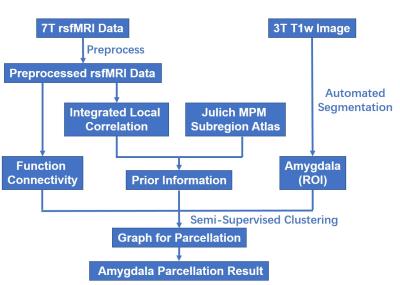
Data processing procedure was shown in Fig.1. The rsfMRI data was preprocessed using SPM12 and CONN, including slice-timing correction, head motion correction, registration of 7T anatomical and functional scans to 3T anatomical image space, and nonlinear registration to MNI standard space, spatially smoothing with 6mm FWHM Gaussian kernel, temporally band-pass filtering (0.01-0.08Hz), regress out nuisance covariates (time series predictors for cerebrospinal fluid, white matter, and six motion parameters). Finally, the rsfMRI data were resampled to have a spatial resolution of 1.5mm.
Functional parcellation of the amygdala
The amygdala of each individual subject was parcellated into 3 subregions using a semi-supervised clustering (SSC) algorithm2.Particularly, a graph was built by modeling each voxel in amygdala as a graph node and connecting each pair of nodes with an edge weighted by a similarity measure of their functional signals. Then, the graph was parcellated into 3 subgraphs by optimizing similarity of nodes within each subgraph and similarity between the parcellation result and prior information that was derived from a cytoarchitectonic maximum probability map (MPM) of amygdala subregion in the SPM Anatomy toolbox. Instead of directly using the MPM representation, we identified one small region with homogeneous functional signals in each subregion as a prior information.
Results
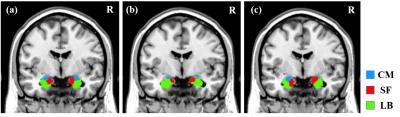
The amygdala of each subject was parcellated into centromedial (CM), laterobasal (LB), superficial (SF) subregion. A maximum probabilistic map (SSC-MPM) of parcellation results for bilateral amygdala obtained from 20 subjects is shown in Fig.2a. As shown in Fig.2b, the parcellation results were different from the cytoarchitectonic MPM, indicating they were not biased to a prior information. The SSC-MPM also highlighted inter-subject variability. The parcellation result of a randomly selected subject is shown in Fig.2c.
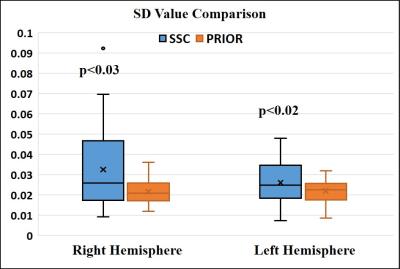
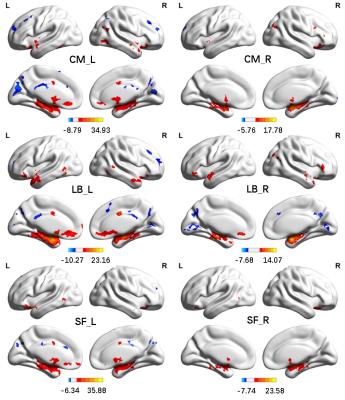
The parcellation results were also evaluated with respect to their functional homogeneity of subregions, measured by modified silhouette distance3 (SD). Results in Fig. 3 showed that the functional parcellation had higher SD than the cytoarchitectonic parcellation (p<0.02 and 0.03 for left and right amygdala respectively), indicating that the functional parcellation had better functional homogeneity. Subregions of the amygdala displayed distinctive FC patterns, as shown in Fig. 4.
Discussion
Based on 7T rsfMRI data, bilateral amygdala was successfully parcellated into 3 subregions. These subregions had distinctive FC patterns, indicating the parcellation results were functionally meaningful. The parcellation results shared similarity with the cytoarchitectonic parcellation albeit different due to a prior information adopted in the parcellation method. Better functional homogeneity of subregions had been achieved, indicating that individualized functional parcellation results of the amygdala better characterize distinctive subregions and capture inter-subject variability.Conclusions
Based on high spatial resolution rsfMRI data at 7T, human amygdala can be parcellated into 3 subregions with distinctive functional connectivity patterns. These individual-level parcellation results might inform personalized medicine and better capture inter-individual variability in neuroimaging studies of the amygdala.Acknowledgements
We acknowledge Ms. Jing An (Siemens Shenzhen MR Ltd., China) and Ms. Yifan Miao (Siemens HealthCare, China) for their technical support. This work was supported in part by National Key Basic Research and Development Program (No. 2015CB856404, No. 2015CB351701), CAS grants (XDB02010001 and XDB02050001), and National Natural Science Foundation of China (No. 81271514, 61473296, 91132302).References
1.Bzdok, Danilo, et al. "An investigation of the structural, connectional, and functional subspecialization in the human amygdala." Human brain mapping34.12 (2013): 3247-3266.
2.Cheng, Hewei, and Yong Fan. "Functional parcellation of the hippocampus by clustering resting state fMRI signals." 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI). IEEE, 2014.
3.Cheng, Hewei, and Yong Fan. "Semi-supervised clustering for parcellating brain regions based on resting state fMRI data." SPIE Medical Imaging. International Society for Optics and Photonics, 2014.
Figures