5244
Segmented EPI reconstruction based on physiological information for fMRI studiesGuoxiang Liu1 and Takashi Ueguchi1
1National Institute of Information and Communications Technology, Suita-shi,Osaka, Japan
Synopsis
We proposed a new segmented EPI reconstruction method based on physiological information to reduce influence of physiological noise. A human brain was scanned on a 7T MRI scanner using segmented EPI, while recording cardiac pulse and respiration data. Our results showed that the proposed reconstruction method can reduce the respiratory-related and cardiac-related signal changes in fMRI studies.
Purpose
Ultrahigh-field (≥7T) high-resolution functional MRI (fMRI) has shown its importance for subcortical areas studies. Although high fields can increase sensitivity to the blood oxygen level-dependent (BOLD) effect and signal-to-noise ratio (SNR), many researchers prefer 3T due to the limited availability of ultrahigh-field scanners and their technological constraints. Segmented echo planar imaging (EPI) is a promising approach for high-resolution fMRI as it provides an increased SNR. However, physiological noise will reduce temporal SNR (tSNR) very much, because different parts of k-space data are acquired in different physiological situations (cardiac pulse, respiration) 1. Some image-based or ICA-based physiological noise correction methods have been reported, but those kinds of post-processing methods may influence the reliability of analyzed results. In this work, we proposed a new segmented EPI reconstruction method based on physiological information to reduce influence of physiological noise. A human brain was scanned on a 7T MRI scanner using segmented EPI, while recording cardiac pulse and respiration data. Our results showed that the proposed reconstruction method can reduce the respiratory-related and cardiac-related signal changes in fMRI studies.Methods
The traditional segmented EPI reconstruction method reconstruct one image by using continuous segmented k-space data, which are acquired in different physiological situation in fMRI studies. Just considering the MR imaging technique, it is possible to reconstruct one image using discontinuous segmented k-space data. The proposed method select the different segmented k-space data acquired in similar physiological situation to reduce the respiratory-related and/or cardiac-related artifact in one image and then increase the tSNR of images series in this physiological situation. If the volume number is big enough, BOLD response can be analyzed in different physiological situation respectively. A human brain was scanned using segmented EPI sequence on a Siemens MAGNETOM 7T scanner with a 32-channel phased array head coil (Nova Medical, MA, USA) to obtain 720 segmented EPI data (144 volumes, segment 5) at spatial resolution of 1×1×1 mm3. Acquisition parameters were as follows: TR = 1000 ms, TE = 25 ms, flip angle = 50°. In order to evaluate how the physiological noise affect analysis in common fMRI studies, a standard block designed visual grating bar task was performed in the experiment with stimulation ON and OFF for blocks of 9 s each, total 18 shots for one ON/OFF unit and repeated 40 times. In other words, there are 18 timing points for detecting BOLD response in one ON/OFF unit, for each timing point, there 40 candidates segmented k-space date in 40 repetitions. And those candidates appeared as different segmented parts, because the length of ON/OFF unit is not integral multiple of number of shot (ON/OFF unit 18 and shot 5). At each timing point, 8 images can be reconstructed. For a given specific physiological situation, we can select segmented parts acquired in this physiological situation to reconstruct an image at this time point. In the same way, all 18 images for one ON/OFF unit can be reconstructed almost without physiological noise (data were acquired in the similar physiological situation). During the scanning, physiological information (cardiac and respiration) was recorded. Three cardiac situations and three respiration situations were used for comparison. BrainVoyagerQX (Version 2.8.4.2645, Brain Innovation, Maastricht, The Netherlands) was used to analyze time series without any preprocessing. Generalized linear model (GLM) analysis was applied with stimulation ON/OFF as a binary regression variable.Results and Discussion
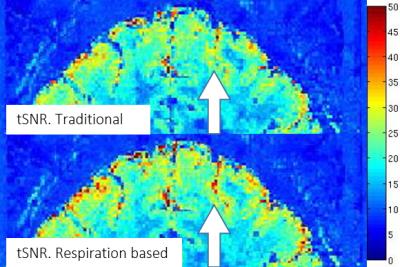
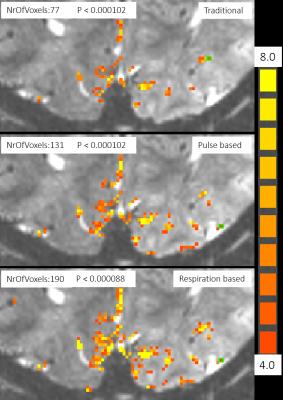
Raw data, recorded physiological information, and acquisition time of each segmented k-space part (recorded in DICOM header) were used to reconstruct images based on cardiac and respiration situations. Compared to traditional reconstructed images, images reconstructed based on respiration situation provided clear tSNR increment shown in Fig. 1. The number of pixels “activated” by GLM analysis using same parameters also show the impact of our proposed method. From the analysis shown in Fig. 2, we found that the most effective way to reduce physiological noise is to reconstruct images based on respiration situation, because respiration noise strongly affected the analysis. One possible reason is that the respiration may cause brain motion and variation of oxygen concentration.