5168
Accelerating T2* Mapping with Maximum Likelihood Estimation (MLE) and Parallel Imaging (PI)1Institute for Digital Communication, University of Edinburgh, Edinburgh, United Kingdom, 2Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, United Kingdom
Synopsis
The utility of MR parametric mapping is limited due to the lengthy acquisition time. A Maximum Likelihood Estimation (MLE) and Parallel Imaging (PI) method is presented for MR parameteric mapping. The approach is based on a high Signal to Noise ratio (SNR) assumption such that the noise can be modelled as Gaussian and estimates the parameters that maximizes the signal from a multichannel coil. The method was tested on a multi-echo gradient-echo T2* mapping experiment in a phantom and a human brain. Accurate T2* maps were reconstructed up to an acceleration factor of 6 with a small error for phantom and human brain.
Background
The estimation of MR parameters such as relaxation times (T1, T2, T2*) requires the acquisition of multiple images at different sequence parameters. Parameter estimation is achieved by fitting the signal evolution with a parameter-dependent model on a pixel-wise basis. It has been demonstrated that the accurate estimation of relaxation times can be done over a wide range of SNR and phased array coil configurations with the MLE technique 1, 2. The aim of this study is to present a method based on MLE that can estimate the relaxation times in conjunction with PI methods.Theory
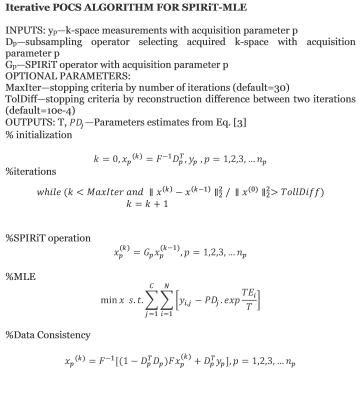
The mono-exponential model applied to decay of a multiecho T2 sequence is given by $$M(TE)_{i}=PD.exp\frac{-TE_{i}}{T}....(1)$$ Here, Pseudo Density(PD) represents the signal amplitude for an echo time TE = 0 and T is the relaxation time. Because the repetition time is fixed, PD is the product of three unknown factors: T1 weighting, PD and receiver coil response 2. Conventionally parameters are estimated by least squares which minimizes the residual sum of squares between observed data and exponential model at the ith TE over N echoes: $$ X^{2}=\sum_{i=1}^{N}[y_{i}-PD.exp\frac{-TE_{i}}{T}]....(2)$$ Multichannel coil is used to enhance SNR3 and to accelerate the image acquisition by employing PI4,5. In the case of images acquired with parallel methods, the distribution of noise in the image is dependent upon the coil sensitivity profiles and varies spatially as well as with the reconstruction method. According to MLE, for a given number of coil channels the best estimate of relaxation time T will be derived by maximizing with respect to PDc and T the joint probability distribution:$$\ln P_{C}(y_{C};PD_{C},T)=\ln \prod_{i=1}^NP\left\{y_{C}(TE_{i});PD_{C},T\right\}=-\sum_{i=1}^C\sum_{j=1}^N\left[y_{i,j}-PD_{i}.exp^{\frac{-TE_{j}}{T}}\right]...(3)$$where $$$P\left\{y_{C}(TE_{i});PD_{C},T\right\}$$$ is the joint signal probability distribution. In the case of higher SNR when the noise distribution can be considered normal, the approximation to the ML is equivalent to the weighted least squares solution where we have also assumed that the variance of noise in case of multichannel coil is uncorrelated across the coils. Our MLE approach estimates the proton density which is weighted according to the sensitivity information of the coils and a relaxation time which maximizes the signal from all the coils according to the equation (3). For parallel imaging, the SPIRiT operator (Gp) with acquisition parameter p that multiplies SPIRiT kernel in image space is used 6. To accelerate the parametric mapping, the problem can be solved using a projection onto convex sets (POCS) algorithm applying MLE and SPIRiT iteratively to undersampled data in order to impose both the exponential relaxation model and the SPIRiT multi-coil model. Details of the algorithm can be seen in Figure(1).
Methods
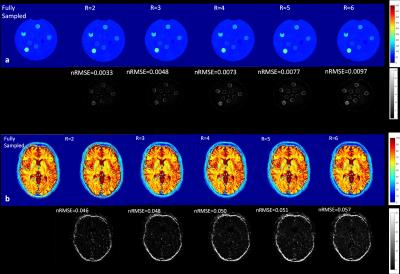
The proposed approach was demonstrated for T2* mapping in the phantom comprising nine compartments (T2* values 65-250 ms). Furthermore, an in-vivo experiment was performed in a healthy human brain. All measurements were performed on a 1.5T clinical scanner (GE Healthcare, Waukesha, WI, USA) using a 3D-enhanced fast gradient-recalled echo sequence with the following parameters(16 echoes, TR = 87 ms, FOV = 256mm, 256x256 matrix, readout bandwidth=31.56 KHz, flip angle 15°, 2mm slice thickness). The total acquisition time for acquiring 32 slices was approximately 18 minutes. The dataset was retrospectively undersampled by factors of 2, 3, 4, 5, and 6 with variable density Poisson disk pattern including an autocalibration region of 24 x 24 in the ky–kz plane. A 7x7 SPIRiT kernel was calibrated from the autocalibration region and the undersampled data was reconstructed by SPIRiT and MLE using proposed POCS reconstruction. To determine the error of the estimated T2* maps, the normalized root mean square error (nRMSE) was calculated using the formula: $$nRMSE=\frac{1}{max(T2)-min(T2)}\sqrt{\frac{1}{N_{i}}\sum_{i=1}^{N_{i}}((T2^*(i)-{T}2^{\prime*}(i))^2}....(4)$$where T2* is the fully sampled dataset, and T2'* is the accelerated map and Ni is the number of image pixels.
Results
Figure 2 (a&b) shows the results for the T2* maps obtained from the fully sampled data and the maps from undersampled data for the phantom and in-vivo measurements, respectively. The number of iterations needed for the reconstruction was between 10 and 15. The nRMSE with respect to the fully sampled map is given below each map. T2* maps from phantom and the volunteer showed the same trend with increased nRMSE with increasing acceleration factor.Conclusion
The proposed MLE-SPIRiT reconstruction allows reconstruction of T2* maps with a small number of iterations. The method allows significant reduction of the required data without compromising the quality of the parameter maps. This method can be a way forward towards the application of MR parameter mapping in clinical scenarios with reduced scan acquisition time.Acknowledgements
The research leading to these results has received funding from the European Union's Seventh Framework Programme (FP7-PEOPLE-2013-ITN) under grant agreement n° 607290 SpaRTaN. The work was carried out on a 1.5 T GE Signa clinical scanner operating within the Brain Research Imaging Center(BRIC), Edinburgh Imaging, University of Edinburgh.References
1. Hardy PA, Andersen AH. Calculating T 2 in Images from a phased array receiver. Magn Reson in Med. 2009;61(4):962-969.
2. Bonny JM, Zanca M, Boire JY, Veyre A. T2 maximum likelihood estimation from multiple spin-echo magnitude images. Magn Reson Med.1996;36:287–293.
3. Wright SM, Wald LL. Theory and application of array coils in MR spectroscopy. NMR Biomed 1997;10:394–410.
4. Pruessmann K, Weiger M, Scheidegger M, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med 1999;42:952–962.
5. Griswold M, Jakob P, Heidemann R, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002;47:1202–1210.
6. Lustig M, Pauly J. SPIRiT: iterative self-consistent parallel imaging reconstruction from arbitrary k-space.Magn ResonMed 2010;64:457–471.
Figures