5162
A new pattern for Autocalibrated Parallel Imaging Reconstruction for GRASE: APIR4GRASE1Departments of Medical Informatics and Radiology, Erasmus MC, Rotterdam, Netherlands, 2Departments of Radiology and Nuclear Medicine, Erasmus MC, Rotterdam, Netherlands, 3Imaging Science and Technology, Delft University of Technology, Delft, Netherlands
Synopsis
We propose a subsampled interleaved parallel acquisition pattern for Autocalibrated Parallel Imaging Reconstruction for GRASE (APIR4GRASE) which considers different echoes during each refocusing of the GRASE as if they originated from different coil channels. APIR4GRASE eliminates ghosting artifacts caused by the phase and amplitude modulations in traditional GRASE split sampling pattern and achieves an additional acceleration factor of 1.3 compared to a fully sampled GRASE k-space. In addition, multiple contrast (spin echo and gradient echo) images are reconstructed. Experiments on a phantom demonstrate the effectiveness of our method.
Purpose
3D GRASE extends 3D Fast/Turbo Spin Echo (FSE/TSE) by sampling spin echoes (SE) with gradient echoes (GRE). Traditionally1, spin echo and gradient echo signals are distributed in separate regions in k-space to reduce artifacts caused by the phase and amplitude modulations. However, some artifacts still remain. Additionally, the large phase-encode (PE) steps between echoes can induce important eddy currents. To address these issues we propose a subsampled interleaved parallel acquisition pattern for Autocalibrated Parallel Imaging Reconstruction2 for GRASE (APIR4GRASE), which in addition provides acceleration compared to a fully sampled GRASE k-space and produces multiple contrast (spin echo and gradient echo) images at the same time.Methods
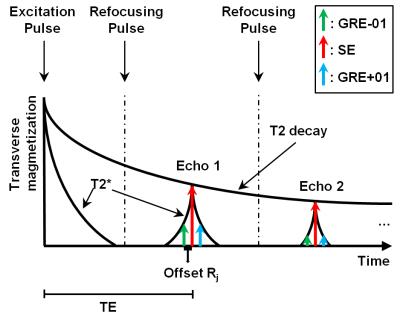
In the 3D GRASE acquisition, during each refocusing of the FSE, multiple echoes are acquired (three in our case: GRE-01, SE, GRE+1). As shown in Figure 1, SE is the T2 weighted echo and GRE samples a T2* decay from the SE.
From a parallel imaging perspective, the differences between the echoes can be modelled as different coil channels: The signal obtained from object $$$M({\bf x}) $$$ in channel $$$c$$$ at time offset $$$R_{j}$$$ of echo $$$j$$$ from the spin echo can be written as $$$S_{j,c}({\bf x})= M({\bf x}) C_{c}({\bf x})e^{-|R_{j}|/T2^{*}-i\omega_{0}({\bf x})R_{j}}$$$, where $$${\bf x}\in\mathbb{R^{3}}$$$ is the location of the signal, $$$C_{c}$$$ is the coil sensitivity of channel $$$c$$$ and $$$\omega_{0}({\bf x})$$$ is the off resonance frequency. If we now assume $$$C'_{j,c}({\bf x})=C_{c}({\bf x})e^{-|R_{j}|/T2^{*}-i\omega_{0}({\bf x})R_{j}}$$$ as the sensitivity of echo $$$j$$$ in channel $$$c$$$, then the signal of echo $$$j$$$ in channel $$$c$$$ can be rewritten as $$$S_{j,c}({\bf x})= M({\bf x}) C_{j,c}({\bf x})$$$. Therefore, we propose APIR4GRASE to include the different echoes in GRASE as different channels. Note that the assumed spatial smoothness of $$$C_{c}$$$ is typically preserved when $$$|R_{j}|\ll T2^{*}$$$.
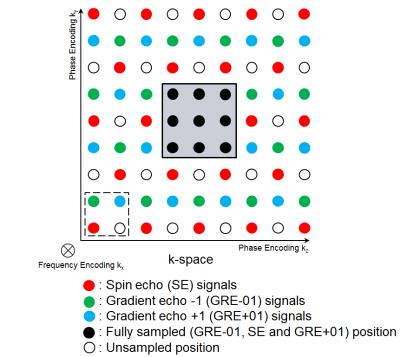
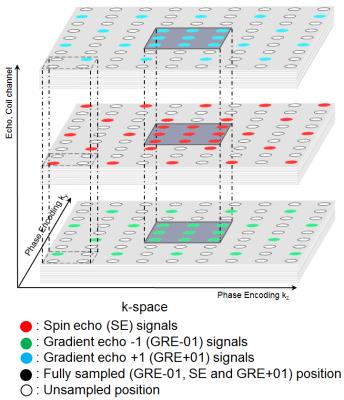
A subsampled interleaved parallel acquisition pattern (Figure 2) is proposed. It is a sheared replication of a 2×2 cell in the two PE dimensions, where in each cell three positions are sampled with SE, GRE-01, GRE+01 respectively. In this pattern, each k-space position is close to every kind of echo. The center part of k-space is fully sampled with all echoes as auto-calibration signal (ACS) region. Different echoes are regarded as coil channels (Figure 3) by APIR4GRASE. It reconstructs full k-spaces for each channel and after Fourier transform reconstructs with sum of square images for each kind of echo. Compared to a split GRASE pattern1, this acquisition reduces eddy current problems by reducing the PE steps between neighboring echoes.
Experiments
A phantom was scanned with the 3D GRASE sequence with EPI factor three on a 3T GE (MR750) scanner using an 8-channel head coil. Full k-spaces for GRE-01, SE and GRE+01 were acquired with the settings of TR/TE=600ms/57.98ms, ETL/ESP=32/8.4ms, matrix=256×256, FOV=220×220mm2, number-of-slices=120, slice-thickness=2mm and BW/pixel=781.25Hz with variable-flip-angle3. Several k-spaces with different subsampling patterns were constructed from the full k-spaces. The proposed sampling pattern and Park’s pattern3 were reconstructed by APIR4GRASE. A prominent difference between the proposed and Park’s pattern is that the latter has no unsampled k-space positions. For comparison a 2x2 subsampled FSE k-space containing only the SE of the proposed pattern was reconstructed by regular ACPI. The size of the ACS in the two PE dimensions was 25×25. Additionally, the split GRASE pattern and the full SE k-space were extracted and reconstructed by inverse Fourier transform. Mean squared error (MSE) to the full SE image was calculated. For each image we evaluated the SNR= mean of signals in a homogeneous region in the water in the phantom/ the noise standard deviation of background region containing artifacts.Results
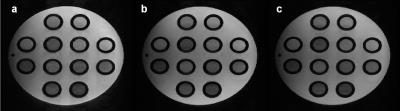
Figure 4 shows the center slice of GRE-01, SE and GRE+01 images reconstructed from our sampling pattern with APIR4GRASE.
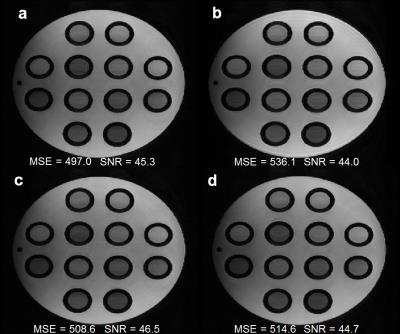
Figure 5 shows that the proposed pattern with APIR4GRASE (5a) eliminates ghosting artifacts present in the split GRASE pattern (5b), while having a better MSE than the subsampled FSE k-space with the regular ACPI (5c) and Park’s pattern (5d) though the SNR is comparable. Additionally, the proposed pattern achieves an acceleration factor of 1.3 compared to Park’s pattern and the split GRASE pattern which are fully sampled.
Discussion and Conclusion
Our method achieves better image quality with additional acceleration compared to the split GRASE acquisition and additionally reconstructs multi-contrast images. Future work includes investigation of different sampling patterns and in-vivo evaluation experiments.Acknowledgements
No acknowledgement found.References
1. Oshio K, Feinberg D A. GRASE (Gradient-and Spin-Echo) imaging: A novel fast MRI technique. Magnetic resonance in medicine, 1991; 20(2): 344-349.
2. Griswold M A, Jakob P M, Heidemann R M, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magnetic resonance in medicine, 2002; 47(6): 1202-1210.
3. Kim H, Kim D H, Park J. Variable-flip-angle single-slab 3D GRASE imaging with phase-independent image reconstruction. Magnetic resonance in medicine, 2015; 73(3): 1041-1052.
Figures