5105
Semi-automated Segmentation of Hip Cartilage in Physiological Magnetic Resonance Imaging: A Fast, Accurate, and Clinically Viable Methodology1Nuffield Department of Orthopaedics, Rheumatology, and Musculoskeletal Sciences, University of Oxford, Oxford, United Kingdom, 2Department of Computer Science, University of Oxford, Oxford, United Kingdom
Synopsis
Physiological Magnetic Resonance imaging (pMRI) offers the potential of diagnosing osteoarthritis at a stage where patients may benefit from intervention, and acting as an assay of disease to test the efficacy of novel early intervention treatments. pMRI data, however, requires segmentation to allow morphological and biochemical quantitative analysis. Manual segmentation is time consuming and a viable automated segmentation method in the hip remains elusive. We have produced a fast, accurate, and reproducible semi-automated method of segmentation to allow wider implementation of pMRI for use in quantitative analysis of early OA in the hip in both research and clinical settings.
Purpose
To use physiological MRI (pMRI) in the diagnosis of early OA requires the anatomical structure of interest, such as the articular cartilage, first be isolated from neighboring structures, i.e. segmented1. However, obtaining accurate segmentations can be problematic2. Manual segmentation is time consuming, expensive, requires months of experience, and has been referred to as perhaps the main obstacle for the translation of promising quantitative MRI techniques into clinical practice3. Several successful semi-automatic and automatic methods have been proposed for cartilage segmentation in the knee2,4-8. In contrast, there have been very few successful examples of semi-automated or automated segmentation in the hip. The Partial Volume Effect (PVE) seen at the close interface of the femoral and acetabular cartilage, caused by the spherical nature of the joint, and thinner cartilage than in the knee9 are likely reasons for this. Previous automated segmentation methods have been described in the hip10-14, however, these methods have lacked clinical viability due to requiring traction or training data, having long processing times, not being widely available, or not providing any biochemical quantitative analysis. We aimed to produce a semi-automated method of segmentation of dGEMRIC scans of the hip that is as accurate and reproducible as manual segmentation, but significantly faster, thus allowing wider clinical and research implementation of pMRI.Methods
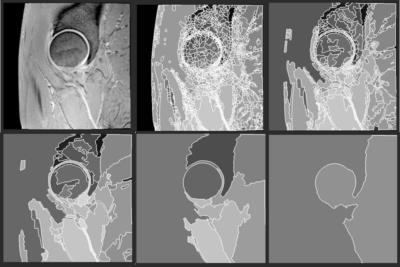
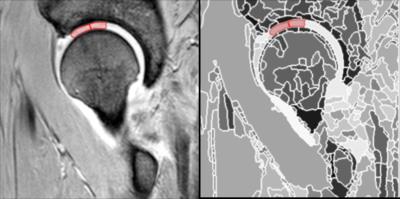
Our semi-automated method consists of a two-step process: a fully automated step which outputs a hierarchical partitioning of the data volume, followed by a user selection of the regions of interest using a region editor. The hierarchical partitioning is generated using a bespoke segmentation approach applied recursively15,16. At each stage of the recursion a coarser layer of regions is generated (Figure 1). A bespoke data structure and associated algorithms have been designed for this purpose, so that the selection of the region of interest (in this case the cartilage) can span across several layers of the hierarchy17. Once the hierarchy is in place, the user proceeds to select regions that align with the cartilage tissue of interest (Figure 2). This was applied to dGEMRIC scans taken from a prospective longitudinal study of individuals considered at high risk of developing osteoarthritis (SibKids18) which were also manually segmented for comparison.Results
Fourteen hips, each comprising 4 or 5 slices, were segmented both manually and using our semi-automated methodology. Processing time for manual segmentation ranged between 90 and 120 minutes per hip. The overall time range for our semi-automatic segmentation method was between 10 and 15 minutes. In the comparison of the semi-automated segmentations with the manual segmentations the accuracy was 0.9886 and DSC was 0.8803. For manual segmentation intra-observer reproducibility showed an accuracy of 0.9992 and DSC of 0.9410, and inter-observer reproducibility showed an accuracy of 0.9987 and DSC 0.9036 respectively. For semi-automated segmentation intra-observer reproducibility showed an accuracy of 0.9971 and DSC of 0.9732, and inter-observer reproducibility showed an accuracy of 0.9996 and DSC of 0.9738 (Table1).Discussion
The overall processing time for our semi automated segmentation method was significantly faster than previously quoted manual segmentation times of 70-100 minutes19, comparable to semi-automated segmentation times in the knee of 10 minutes4, and considerably quicker than automated segmentation in the hip of 3 and 18 hours2,13. Our level of accuracy is comparable to previous reports where DSC for cartilage was 0.82413. To the authors’ knowledge, this will be the first automated or semi-automated methodology for segmentation of the hip that is intended to be licensed for widespread use. The potential ramifications of this work are two fold. In research, this rapid method of hip segmentation would greatly increase the viability, and reduce the cost, of large-scale trials using pMRI. Clinically, the increased applicability of pMRI may allow its increased use in helping to select appropriate candidates for hip arthroscopy and hip preservation surgery, something that remains a major problem in a procedure that is increasingly performed each year20.Conclusion
This study has proposed a fast, accurate, and reproducible semi automated method for segmentation of physiological MRI in the hip. This is the first such method in the hip that shows clinical applicability and forwards the argument for the widespread use of pMRI as an accurate and efficient clinical tool to select appropriate candidates for hip arthroscopy and hip preservation surgery, and as a research tool capable of large-scale use.Acknowledgements
No acknowledgement found.References
1. Eckstein F, Cicuttini F, Raynauld JP, Waterton JC, Peterfy C. Magnetic resonance imaging (MRI) of articular cartilage in knee osteoarthritis (OA): morphological assessment. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2006;14 Suppl A:A46-75.
2. Fripp J, Crozier S, Warfield SK, Ourselin S. Automatic segmentation and quantitative analysis of the articular cartilages from magnetic resonance images of the knee. IEEE transactions on medical imaging. 2010;29(1):55-64.
3. Pedoia V, Majumdar S, Link TM.
Segmentation of joint and musculoskeletal tissue in the study of arthritis.
Magma. 2016;29(2):207-21.
4. Folkesson J, Dam EB, Olsen OF,
Pettersen PC, Christiansen C. Segmenting articular cartilage automatically
using a voxel classification approach. IEEE transactions on medical imaging.
2007;26(1):106-15.
5. Prasoon A, Igel C, Loog M, Lauze F, Dam
EB, Nielsen M. Femoral cartilage segmentation in knee MRI scans using two stage
voxel classification. Conference proceedings :
Annual International Conference of the IEEE Engineering in Medicine and
Biology Society IEEE Engineering in Medicine and Biology Society Annual
Conference. 2013;2013:5469-72.
6. Carballido-Gamio J, Majumdar S. Atlas-based
knee cartilage assessment. Magnetic resonance in medicine. 2011;66(2):574-83.
7. Shan L, Zach C, Charles C, Niethammer
M. Automatic atlas-based three-label cartilage segmentation from MR knee
images. Medical image analysis. 2014;18(7):1233-46.
8. Dodin P, Pelletier JP, Martel-Pelletier
J, Abram F. Automatic human knee cartilage segmentation from 3D magnetic
resonance images. IEEE transactions on bio-medical engineering. 2010;57(11).
9. Cheng Y, Guo C, Wang Y, Bai J, Tamura
S. Accuracy limits for the thickness measurement of the hip joint cartilage in
3-D MR images: simulation and validation. IEEE transactions on bio-medical
engineering. 2013;60(2):517-33.
10. Nakanishi K, Tanaka H, Sugano N, Sato Y,
Ueguchi T, Kubota T, et al. MR-based three-dimensional presentation of
cartilage thickness in the femoral head. European radiology.
2001;11(11):2178-83.
11. Nishii T, Sugano N, Sato Y, Tanaka H,
Miki H, Yoshikawa H. Three-dimensional distribution of acetabular cartilage
thickness in patients with hip dysplasia: a fully automated computational
analysis of MR imaging. Osteoarthritis and cartilage / OARS, Osteoarthritis
Research Society. 2004;12(8):650-7.
12. Li W, Abram F, Beaudoin G, Berthiaume MJ,
Pelletier JP, Martel-Pelletier J. Human hip joint cartilage: MRI quantitative
thickness and volume measurements discriminating acetabulum and femoral head.
IEEE transactions on bio-medical engineering. 2008;55(12):2731-40.
13. Siversson C, Akhondi-Asl A, Bixby S, Kim
YJ, Warfield SK. Three-dimensional hip cartilage quality assessment of
morphology and dGEMRIC by planar maps and automated segmentation.
Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society.
2014;22(10):1511-5.
14. Xia Y, Chandra SS, Engstrom C, Strudwick MW, Crozier S, Fripp J. Automatic hip cartilage segmentation from 3D MR images using arc-weighted graph searching. Physics in medicine and biology. 2014;59(23):7245-66.
15. Beucher S. Watershed, hierarchical
segmentation and waterfall algorithm. Mathematical morphology and its
applications to image processing Springer Netherlands,. 1994:69-76.
16. Marcotegu B BS. Fast Implementation of
Waterfall Based on Graphs. In Mathematical Morphology: 40 Years On Springer
Netherlands. 2005.
17. Golodetz SM NC, Voiculescu ID, Cameron
SA. Two tree-based methods for the waterfall. Pattern Recognition.
2014;47(10):3276-92.
18. Pollard TC, Batra RN, Judge A, Watkins B,
McNally EG, Gill HS, et al. Genetic predisposition to the presence and 5-year
clinical progression of hip osteoarthritis. Osteoarthritis and cartilage /
OARS, Osteoarthritis Research Society. 2012;20(5):368-75.
19. Pollard TC, McNally EG, Wilson DC, Wilson
DR, Madler B, Watson M, et al. Localized cartilage assessment with
three-dimensional dGEMRIC in asymptomatic hips with normal morphology and cam
deformity. The Journal of bone and joint surgery American volume.
2010;92(15):2557-69.
20. AJR Palmer TM, L Broomfield, J Holton, L Majkowski, GER Thomas, A Taylor, AJ Andrade, G Collins, K Watson, AJ Carr, S Glyn-Jones. Past and projected temporal trends in arthroscopic hip surgery in England between 2002 and 2013. BMJ Open Sport & Exercise medicine. 2016;2:82-7.
Figures