5074
Texture Analysis for Evaluating Image Registration1Electrical Engineering, Geroge Mason University, Fairfax, VA, United States, 2Bioengineering, George Mason University
Synopsis
Accurate image registration is essential for both cross-sectional and longitudinal MR studies. In longitudinal studies aligning same contrast intra-subject images, which are the focus of our work, registration is assumed to be a rigid body problem. This assumption is questionable due to global and local changes in brain volume either due to hydration or atrophy. Consequently, misregistration at the voxel level may occur in these studies, which might lead to subject data being discarded. This misregistration is evident particularly around the cortex. Visual inspection of images is used to determine registration accuracy. While this approach is suitable for assessing alignment of landmark structures, it fails to capture the millimteric or sub-millimetric misregistrations. Automatic metrics can precisely estimate the overall performance of a given registration algorithm by employing an evaluation database. However, to estimate the registration accuracy of a given pair of images, such metrics are unsuitable. In this work we propose texture analysis of a subtraction image to evaluate the registration accuracy of a given pair of same contrast, intra-subject images. Once registered, images are intensity normalized, blurred and subtracted. In the event of registration errors, or violation of the rigid body assumption, the subtraction images have artifacts. The texture features of these artifacts are different from the artifact-free (clean) areas of the subtraction images. Using a texture-based classifier, artifact areas in the subtraction images that indicate failed registration are identified. In addition to determining if the registration has failed, our approach can identify the specific locations of misregistration, which can be corrected, leading to a more inclusive subject data.
Purpose
Accurate image registration is essential for
both cross-sectional and longitudinal MR studies. In longitudinal studies
aligning same contrast intra-subject images, which are the focus of our work,
registration is assumed to be a rigid body problem. This assumption is
questionable due to global and local changes in brain volume either due to
hydration or atrophy1,2. Consequently,
misregistration at the voxel level may occur in these studies, which might lead
to subject data being discarded3. This
misregistration is evident particularly around the cortex. Visual inspection of
images is used to determine registration accuracy. While this approach is
suitable for assessing alignment of landmark structures, it fails to capture
the millimteric or sub-millimetric misregistrations. Automatic metrics can precisely
estimate the overall performance of a given registration algorithm by employing
an evaluation database4. However, to
estimate the registration accuracy of a given pair of images, such metrics are unsuitable.
In this work we propose texture analysis of a subtraction image to evaluate the
registration accuracy of a given pair of same contrast, intra-subject images.
Once registered, images are intensity normalized, blurred and subtracted. In
the event of registration errors, or violation of the rigid body assumption,
the subtraction images have artifacts. The texture features of these artifacts
are different from the artifact-free (clean) areas of the subtraction images. Using
a texture-based classifier, artifact areas in the subtraction images that
indicate failed registration are identified. In addition to determining if the
registration has failed, our approach can identify the specific locations of
misregistration, which can be corrected, leading to a more inclusive subject
data.Methods
T2-weighted dual echo TSE sequence were acquired for 20 patients over a year (14 women, 4 men, and mean age 33.6 ± 6.9). T2-weighted image at time-0 (T0) was used as baseline and T2-weighted image at time-11 (T11) (current image) for all the subjects were registered to the baseline image. Following registration, brain extraction, intensity normalization, Gaussian blurring and digital subtraction were performed to generate subtraction images. Eleven texture features5 were generated from the twenty subtraction images. To maintain uniformity, the slice representing the start of bi-lateral ventricles was selected for texture feature testing. Among the twenty subjects, five subjects were selected at random for training the classifier. A threshold based classifier was designed by generating eleven histograms (one for each feature) of the five training images, estimating the threshold using the Otsu method per feature6. These thresholds were averaged across five subjects to estimate eleven global thresholds. The global thresholds were then applied on the texture feature maps of the fifteen remaining subjects. In order to validate the classifier, manual marking of the artifact area was created from a testing example. The Dice-Sorenson co-efficient7(DSC) was estimated between the manually delineated artifact area and the artifact area segmented by the texture feature classifier, were DSC>0.7 is treated as excellent agreement8. A receiver operating characteristic (ROC) curve was generated by varying the threshold to evaluate the performance of the classifier in labeling a voxel as an artifact. To determine if the registration was successful, we computed the artifact area segmented by the texture feature classifier and artifact areas above a certain threshold indicated failed registration. The registration was visually inspected and labeled as failed or successful registration. A receiver operating characteristic curve was generated by varying the area threshold for the artifact area to evaluate the performance of our approach.Results
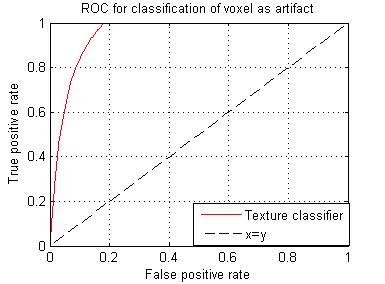
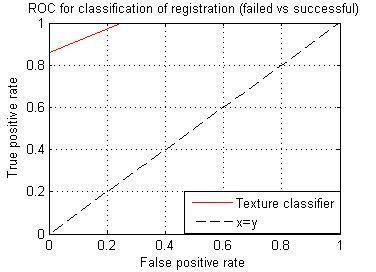
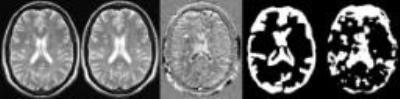
Failed registration leads to artifacts, these artifacts represent inhomogeneity in subtraction images. Among the eleven texture features generated, we selected a feature with the highest DSC (0.7242). The images in figure 1, represent steps involved in classification using the texture feature classifier. Figure 2, shows the ROC for the texture classifier in labeling a voxel as artifacts. Figure 3, shows the ROC for the texture classifier in labeling a given set of images as aligned correctly or failed registration.Conclusion
We presented an automatic technique for evaluating registration accuracy. This technique uses texture analysis to measure the extent of artifacts on subtraction images, which are generated after registration. This approach can indicate failures of registration at millimetric precision. Based on the ROC curve analysis, it is evident that the texture classifier is accurate in labeling a voxel as artifact and labeling a set of images as misregistered. It can potentially benefit multiple studies that rely on accurate image registration.Acknowledgements
No acknowledgement found.References
[1].Duning, T. et al. Dehydration confounds the assessment of brain atrophy. Neurology 64, 548–550 (2005). [2]. Streitbürger, D.-P. et al. Investigating Structural Brain Changes of Dehydration Using Voxel-Based Morphometry. PLOS ONE 7, e44195 (2012).[3]. Duan, Y. et al. Segmentation of subtraction images for the measurement of lesion change in multiple sclerosis. AJNR Am. J. Neuroradiol. 29, 340–346 (2008). [4]. Christensen, G. E. et al. in SpringerLink 128–135 (Springer Berlin Heidelberg). [5]. Haralick, R. M., Shanmugam, K. & Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. SMC-3, 610–621 (1973). [6]. Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979). [7]. Dice, L. R. Measures of the Amount of Ecologic Association Between Species. Ecology 26, 297–302 (1945). [8]. Zijdenbos, A. P., Dawant, B. M., Margolin, R. A. & Palmer, A. C. Morphometric analysis of white matter lesions in MR images: method and validation. IEEE Trans. Med. Imaging 13, 716–724 (1994).