5070
Improved Denoising of Dynamic Arterial Spin Labeling with Infimal Convolution of Total Generalized Variation Functionals (ICTGV)1Institute of Medical Engineering, Graz University of Technology, Graz, Austria, 2Institute of Mathematics and Scientific Computing, University of Graz, Austria
Synopsis
Dynamic arterial spin labeling MRI provides important quantitative information about blood arrival time and perfusion. However, the inherently low signal-to-noise ratio requires repeated measurements to achieve a reasonable image quality. This leads to long acquisition times and hence increases the risk of motion artifacts, which impedes clinical applicability. To overcome this limitation we propose to reconstruct the dynamic ASL data employing ICTGV regularization from a reduced number of averages. The performance of the method is evaluated on synthetic and in-vivo ASL data.
INTRODUCTION
Dynamic arterial spin labeling MRI provides important quantitative information about blood arrival time and perfusion, by measuring the difference between control (C ) and spin-labeled (L) data for multiple post-labeling delays. However, due to an inherently low signal-to-noise ratio of the difference between C and L, repeated measurements are required to achieve a reasonable image quality. This leads to long acquisition times and hence increases the risk of motion artifacts, which impedes clinical applicability. To overcome this limitation we propose to reconstruct the dynamic ASL data employing ICTGV regularization1 from a reduced number of averages. ICTGV proved to be a suitable regularization functional to stabilize reconstruction from highly undersampled dynamic MRI applications and is therefore expected to allow a suppression of noise in dynamic ASL. The performance of the proposed approach is evaluated on synthetic and in-vivo ASL data.METHODS
For baseline evaluation a numerical phantom was generated based on a high-resolution T1w-MRI of our database. The phantom was computed using Matlab and SPM12 as described in Zhao et al.2 MRI rawdata was simulated with 20 artificial receiver-coil sensitivities and different noise-levels. In-vivo ASL data was acquired from one subject using FAIR-QII with a 3D-GRASE readout.3 The following acquisition parameters were used on a clinical 3T system: TR/TE = 2500/20ms, matrix 64x64, voxel size 3x3x3mm³, 30 slices, 6 segments, Turbo-factor = 15, EPI-factor = 21, 9 post-labeling delays: TI1,1 / TI1,2 / TI1,{3-9} = 300 / 600 / 800, TIfirst/ ΔTI /TIlast = 500 / 250 / 2500 ms. 7 averages for each TI were acquired resulting in a total acquisition time of approximately 33min. Reconstructions were computed with ICTGV regularization usging the AVIONIC framework (https://github.com/IMTtugraz/AVIONIC), from the different noise-levels of the synthetic phantom as well as for seven to one averages from in-vivo data.RESULTS
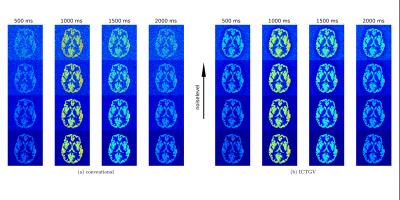
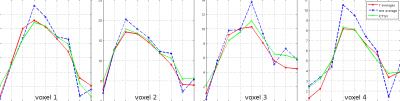
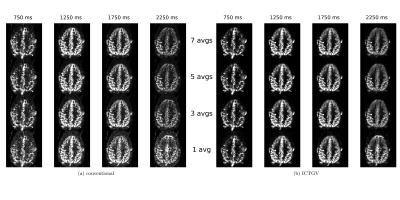
Figure 1 displays the recovery of noisy phantom data by means of ICTGV reconstruction for realistic, linearly increasing noise levels for different post-labeling delays. Figure 2 shows results from invivo data, similar as in Figure 1, but with recovery from different amounts of averages avg=(7,5,3,1) comparing to non-denoised data. The corresponding series of all post-labeling delays for selected voxels comparing gold-standard 7fold averaging, non-averaged ICTGV reconstruction and conventional reconstruction, resp., is displayed in Figure 3.DISCUSSION AND CONCLUSION
The results
demonstrate that a substantial suppression of noise can be achieved
using the proposed regularization approach. This is accomplished by
taking into account correlations of the varying signal intensity
along the marked post-labeling delays. Image quality can be achieved with ICTGV reconstruction from non-averaged
data, as with seven-fold averaging.
This reduces the scanning time from 33min to 4min 45s.
This improvement potentially helps to enable the
consolidation of clinical applicability and higher spatial
resolutions. Further improvements with this method may be achieved by
computing reconstructions from the 3D-parametric data, which was
processed slice-by-slice in this work to reduce the
computational burden. The implementation as well as comparison to
state-of-the-art methods will be subject to future work.Acknowledgements
BioTechMed-Graz
Funded by the Austrian Science Fund (FWF) SFB-F3209-18
NVIDIA Corporation Hardware grant support
References
1.
Schloegl M, Holler M, Bredies K, et al. Infimal convolution of total
generalized variation functionals for dynamic MRI. Magnetic Resonance
in Med. 2016; doi:10.1002/mrm.26352
2. Zhao L, Fiedlden SW, Feng X, et al. Rapid 3D dynamic arterial spin labeling with a sparse model-based image reconstruction. Neuroimage 2015;121:205-216
3. Günther M, Oshio K, Feinberg DA. Single-shot 3D imaging techniques improve arterial spin labeling perfusion measurements. Magn Reson Med. 2005;54(2):491-498
Figures