5061
Semi-automated identification of Substantia Nigra in healthy controls and patients with Parkinson's Disease: a feasibility study using MP2RAGE1Neuroimaging Unit, Institute of Bioimaging and Molecular Physiology (IBFM-CNR), Catanzaro, Italy, 2Institute of Neurology, University Magna Graecia, Catanzaro, Italy, 3Healthcare Sector IM&WS S, Siemens Schweiz AG, Renens, Switzerland, 4Institute of Neuroradiology, University Magna Graecia, Catanzaro, Italy
Synopsis
Reliable in vivo assessment of human substantia nigra (SN) requires highly trained operators and different MRI sequences. Advanced techniques have recently facilitated SN identification, but their acquisition in routine clinical practice may not be feasible. MP2RAGE allows for quantitative T1 mapping with an acceptable acquisition time (< 10 minutes). Moreover, SN can be seen on T1 maps, but not on standard MPRAGE. In this study, we tested the feasibility of semi-automated SN identification on MP2RAGE-derived T1 maps by using a thresholding approach, and compared SN volume and T1 values between healthy controls and patients with Parkinson's disease.
Introduction
Reliable identification of brain structures on MRI enables the assessment of regional metrics reflecting complementary tissue properties. Identification of the substantia nigra (SN), site of dopaminergic neurons that project to the striatum, requires the simultaneous observation of multiple MRI sequences from highly trained operators, since its boundaries are not easily detectable on commonly acquired contrasts. This is not only cumbersome, but also involves acquisition of advanced protocols, which is not always feasible in clinical environments. Degeneration of SN neurons, hallmark of Parkinson's disease (PD), has widely occurred when motor symptoms arise. It is therefore crucial to assess its integrity in vivo, searching for early biomarkers of PD.1 Novel sequences have been recently used to characterize SN (e.g., neuromelanin and susceptibility-weighted imaging).1 While facilitating SN identification, these acquisitions are not easily transferable into clinical practice, especially since patients with PD may suffer from resting tremor, which could in turn cause significant motion artifacts during long scans. The Magnetization Prepared 2 Rapid Acquisition Gradient Echoes sequence (MP2RAGE)2,3 has become increasingly popular since it allows, in less than 10 minutes, reconstruction of quantitative T1 maps and of bias-field-independent images with increased grey/white matter contrast. SN is composed by neuronal bodies that lie in the midbrain, surrounded by white matter bundles: since the T1 is inversely proportional to the protein density per volume unit, healthy SN should appear hyperintense compared to surrounding tissue, which is rich in protein content (e.g., myelin). On the contrary, death of SN neurons should be accompanied by protein accumulation (e.g., α-synuclein), thus inducing reductions in T1 values. For all the above-mentioned reasons, aim of this study was to test the feasibility of SN identification, at single-subject level, on quantitative T1 maps. Furthermore, we compared T1 characteristics in healthy controls and patients with PD.Methods
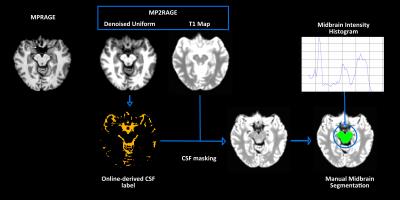
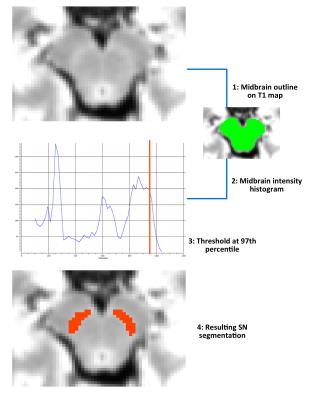
Twelve healthy subjects (7 female, age range 29-42) and 12 PD patients (5 female, age range 56-69) underwent the same 3T MRI protocol (Biograph mMR, Siemens Healthcare), including MPRAGE (TR/TE/TI/TE/TR=900/2.34/2300 ms, flip angle=8°, isotropic voxel size 1 mm) and MP2RAGE sequences (TI1/TI2/TE/TR=700/2500/1.9/5000 ms; flip angles 4° and 5°, isotropic voxel size 1 mm). MP2RAGE data acquisition and online image reconstruction were performed with a vendor-supplied package (WIP #900B-VE11A). For each subject, the following images were used: a uniform denoised MP2RAGE, a quantitative T1 map, labels for subcortical structures and cerebrospinal fluid (CSF). Since all these images are inherently coregistered, the standard MPRAGE was linearly transformed in the MP2RAGE space. Subsequently, semi-automated SN segmentation was performed as shown in Figure 1: first, CSF masking was performed; second, midbrain was manually outlined on the CSF-masked T1 maps, selecting axial slices were SN was more clearly visible; third, intensity histogram thresholding of the midbrain T1 map was performed to identify the percentile that allowed optimal SN segmentation. Of note, midbrain outlining is largely leaner than SN segmentation. In order to assess segmentation accuracy, the intraclass correlation coefficient (ICC) was calculated between thresholding-obtained and manual segmentations of SN performed on the T1 maps by a trained physician. For group comparison, analysis of covariance was performed on SN volume and average T1 value. Since PD patients were older than healthy volunteers, age and sex were included as covariates. Significance threshold was set at 0.05 after Bonferroni correction for multiple comparisons.Results
SN was successfully identified using the semi-automated procedure on all subjects. Histogram analysis revealed that the 97th percentile of the midbrain intensity distribution was the optimal threshold to correctly identify SN on individual T1 maps (Figure 2). The ICC between the manual and semi-automated segmentations was 0.88. Patients with PD showed significantly lower volume and quantitative T1 value compared to controls.Discussion
Our findings suggest that MP2RAGE might become a useful tool for lean identification and quantitative characterization of SN. The relatively short acquisition time of this sequence makes it suitable for routine clinical applications, even in patients with resting tremor, which may cause severe motion artifacts. We found significant reduction of SN volume and averageT1 in patients with PD compared to healthy subjects. Since T1 is inversely proportional to protein density in tissues, decrease in its value might be a marker of neuronal death, which may cause protein accumulation. Acquisition on larger groups of patients and age-matched healthy controls is needed to validate and to possibly fully-automatize SN quantification.Conclusions
Quantitative T1 mapping with MP2RAGE seems promising for SN measurements in patients with PD, not only in research environments but also in routine clinical practice. Assessing SN on large samples of patients could provide a novel marker for early PD diagnosis.Acknowledgements
The authors would like to thank Stefania Randisi for onsite support in installing the WIP software, and Domenico Gullà for acquisition support.References
1. Lehéricy S, Sharman MA, Santos CLD, et al. Magnetic resonance imaging of the substantia nigra in Parkinson's disease. Movement disorders. 2012;27(7):822-830.
2. Marques JP, Kober T, Krueger G, et al. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage. 2010;49:1271-1281.
3. Fujimoto K, Polimeni JR, van der Kouwe AJW, et al. Quantitative comparison of cortical surface reconstructions from MP2RAGE and multi-echo MPRAGE data at 3 and 7 T. Neuroimage. 2014;90:60-73.
Figures