4994
Automated assessment of paraspinal muscles fat composition based on the segmentation of chemical shift encoding-based water/fat-separated images1Department of Diagnostic and Interventional Radiology, Klinikum rechts der Isar, Technical University of Munich, Munich, Germany, 2Department of Diagnostic and Interventional Neuroradiology, Klinikum rechts der Isar, Technical University of Munich, Munich, Germany, 3Philips Research Laboratories, Hamburg, Germany
Synopsis
Chemical shift encoding-based water-fat MRI derived proton density fat fraction (PDFF) of the paraspinal muscles has been emerging as important surrogate marker in subjects with intervertebral disc disease, osteoporosis, sarcopenia, and neuromuscular disorders. However, measurements of paraspinal muscle PDFF are currently limited in clinical routine due to the required time-consuming manual segmentation procedure. The present study aimed to develop an automatic segmentation algorithm of the paraspinal muscles at the lumbar spine based on water-fat MRI and compared the performance of this algorithm to ground truth data based on manual segmentation.
Purpose
MR-based assessment of the paraspinal muscles fat composition has been proposed as surrogate marker in subjects with intervertebral disc disease, osteoporosis, sarcopenia, and neuromuscular disorders1,2. Based on chemical shift encoding-based water-fat MRI, the proton density fat fraction (PDFF) of each paraspinal muscle compartment could be reliably extracted3. For clinical routine, water-fat MRI to assess paraspinal muscle PDFF is currently limited due to the time-consuming manual segmentation procedure. Therefore, the purpose of this study was to develop an automatic segmentation algorithm of the paraspinal muscles based on chemical shift encoding-based water-fat MRI and compare the performance of this algorithm to ground truth data based on manual segmentation.Methods
Subjects: Ten healthy subjects (eight males, two females; age: 29±8 years, BMI: 26.7±2.3 kg/m²) were recruited for this study. All subjects were scanned at baseline and 6-week follow-up.
MR Imaging: The lumbar musculature of the subjects was scanned on a 3 T whole-body scanner (Ingenia, Philips Healthcare, Best, Netherlands) using the built-in-the-table posterior coil elements (12-channel array). An axially-prescribed six-echo 3D spoiled gradient echo sequence was used for chemical shift encoding-based water-fat separation. The sequence acquired the six echoes in a single TR using non-flyback (bipolar) read-out gradients and the following imaging parameters: TR/TEmin/ΔTE = 11/1.04/0.8 ms, FOV = 220x220x219 mm3, acquisition matrix = 72x110x73, acquisition voxel size = 3.1x2.0x3.0 mm3, frequency encoding direction = L/R, fold over suppression = 80 mm (P direction), receiver bandwidth = 2756 Hz/pixel, no SENSE, Navg = 1, scan time = 2min and 1 s per stack. A flip angle of 3° was used to minimize T1-bias effects4.
Imaging-Based Fat Quantification: The gradient echo imaging data were processed on-line using the mDIXON Quant method provided by the manufacturer. It performs a complex-based water-fat decomposition using a pre-calibrated seven-peak fat spectrum and a single T2* to model the signal variation with echo time. PDFF maps were then computed as the ratio of the fat signal over the sum of fat and water signals.
Manual Segmentation: Manual segmentation of the paraspinal muscles was performed on the PDFF maps at baseline and follow-up by using the free open-source software MITK. The following six muscle compartments were separately segmented from the upper endplate level of L2 to the lower endplate level of L5: right and left psoas muscles, right and left quadratus lumborum muscles, and right and left erector spinae muscles (Fig. 1).
Automatic Segmentation: Baseline and follow-up images and corresponding manual muscle segmentations of seven subjects were used as training dataset and those of the remaining three subjects as test dataset. Manual muscle segmentations in the baseline and follow-up images of the three test subjects served as ground truth and were considered as gold standard for the automatic muscle segmentation results. Based on the manual segmentations of the training set, a model of the six muscle compartments was generated. It comprised an average shape model, a dual feature model, associating each surface point with a fat and water image appearance feature, and a detection model. Automatic segmentation first performed a Generalized Hough-Transform for structure localization to initialize the model in the patient image, followed by a coarse to fine individualization of the surface model5 (Fig. 2).
Statistical Analysis: Dice coefficients were determined to compare the automatic muscle segmentations with the corresponding ground truth. Wilcoxon signed rank tests were used to assess differences of muscle volume and PDFF based on automatic segmentation and ground truth, respectively.
Results
The Dice coefficient averaged over all six muscle compartments amounted to 0.83 (min: 0.75, max: 0.90). Table 1 shows the Dice coefficients separately for each muscle compartment. Mean volume and PDFF of each muscle compartment for the training dataset are listed in Table 2. The automatic segmentation algorithm significantly (p<0.05) overestimated the muscle volumes of erector spinae and psoas muscles (Table 2). Differences in PDFF values based on the automatic muscle segmentation as compared to the ground truth were relatively small (range: 0.02 to 0.58%), but statistically significant (p<0.05) in the erector spinae muscles (Table 2).Discussion & Conclusion
The proposed algorithm for automatic paraspinal muscle segmentation on chemical shift encoding-based water-fat MRI showed adequate Dice coefficients and small errors in PDFF (range: 0.02 to 0.58%) in the scanned healthy subjects. Improvement of the algorithm including an increased number of subjects for the training dataset is necessary to reliably extract muscle volume and PDFF of the erector spinae muscles in the future, especially when applying the method in atrophied muscles affected by pathology.Acknowledgements
The present work was supported by Philips Healthcare (to D.C.K.), the European Research Council ERC-StG-2015 677661 (to D.C.K.) and ERC-StG-2014 637164 (to J.S.K.).References
1 Teichtahl AJ, Urquhart DM, Wang Y, et al. Lumbar disc degeneration is associated with modic change and high paraspinal fat content - a 3.0T magnetic resonance imaging study, BMC Musculoskelet Disord 2016;17:439.
2 Dahlqvist JR, Vissing CR, Thomsen C, Severe paraspinal muscle involvement in facioscapulohumeral muscular dystrophy. Neurology 2014;83:1178.
3 Hu HH, Kan HE. Quantitative proton MR techniques for measuring fat. NMR Biomed 2013;26:1609.
4 Karampinos DC, Yu H, Shimakawa A, et al. T1-corrected fat quantification using chemical shift-based water/fat separation: application to skeletal muscle. Magn Reson Med 2011; 66:1312.
5 Buerger C, Peters J, Waechter-Stehle I, et al. Multi-modal Vertebra Segmentation from MR Dixon for Hybrid Whole-Body PET/MR. Computational Methods and Clinical Applications for Spine Imaging, Lecture Notes in Computational Vision and Biomechanics 17, Springer, 2014, p. 159-171.
Figures
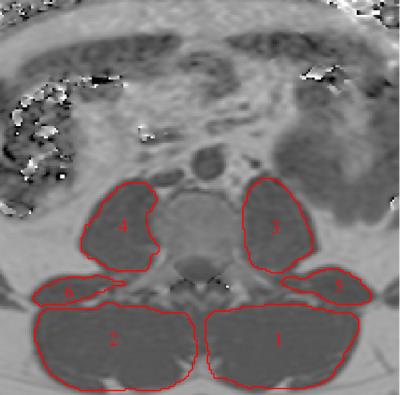
Figure 1:
Representative fat fraction map with manually segmented muscle compartments. 1: left erector spinae muscles, 2: right erector spine muscles, 3: left psoas muscle, 4: right psoas muscle, 5: left quadratus lumborum muscle, 6: right quadratus lumborum muscle.
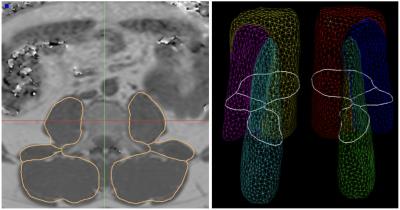
Figure 2:
Left: Result of the automatic segmentation of the muscle compartments (same case as shown in Fig. 1). Right: Average triangular surface model with cross- sectional cut-contour of central axial slice depicted in white.
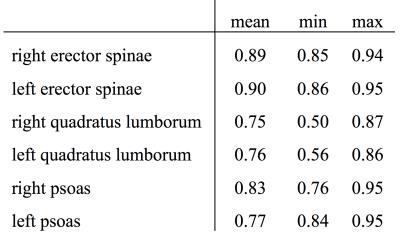
Table 1:
Dice coefficients for each muscle compartment.
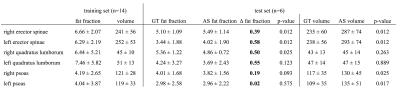
Table 2:
Mean and standard deviation of PDFF [%] and volume [cm³] of each muscle compartment in the training and test dataset. P-values refer to the comparison of Automatic Segmentation (AS) and Ground Truth (GT) results in the test dataset.