4940
Co-registration of Breast MRI and CT Using Gravity Unloading1Tu & Yuen Center for Functional Onco-Imaging, University of California, Irvine, Irvine, CA, United States, 2E-Da Hospital and I-Shou University, Kaohsiung, Taiwan, 3Tzu-Chi General Hospital, Taichung, Taiwan
Synopsis
A biomechanical simulation method for co-registration of breast MRI and low-dose chest computed tomography (LDCT) images is presented, by aligning the images in a virtually unloaded configuration. The breast tissue was considered as neo-Hookean material, and the finite element method was applied to simulate the deformation from gravity-unloading. The Demon’s non-rigid registration algorithm was applied to co-register the gravity-unloaded MRI and LDCT models. Fourteen normal subjects who received both breast MRI and LDCT for breast and lung cancer screening were analyzed. The results show that the pre-processing using gravity unloading can facilitate the co-registration of LDCT and MRI.
Introduction
The standard
treatment for breast cancer includes surgery, with radiotherapy when the
patient receives lumpectomy. The pre-operative MR images contain valuable
information about the size and position of tumor, with the patient imaged in
the prone position. The surgery and
radiotherapy are performed with the patient lying in the supine position. Therefore,
prone-to supine breast image registration may play an important role in image-guided
therapy. Breast is a soft organ, and deforms a lot under different mechanical
loading conditions, e.g. different gravity and body support when the women are
in different body positions, which poses a great challenge for breast image
registration. Conventional registration methods cannot lead to reasonable solutions.
In this study, a finite element method
based image registration, with gravity unloading, was applied to register the
MR images (prone position) and LDCT images (supine position) acquired from the
same patients receiving MRI and CT for breast and lung cancer screening.Method
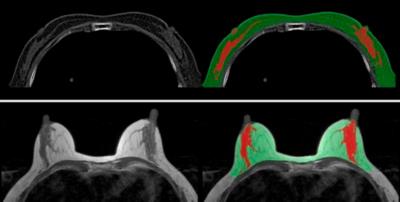
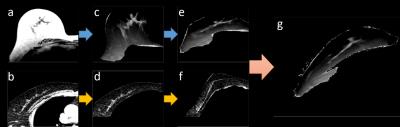
Due to the different imaging modalities, the resolution and the noise level is totally different. First, we applied a Gaussian filter with window size 5 to denoise the LDCT images. Then the MR and LDCT images were adjusted to the same voxel size, and the intensities of MR images were normalized to the same scale as LDCT images, as shown in Fig. 1. A standardized segmentation method based on fuzzy-C-means (FCM) algorithm was applied to LDCT and MR images to segment the breast from the body and segment the fibroglandular tissue1,2, as shown in Fig. 2. Using the segmented results, two 3D breast models can be reconstructed, one for supine position (LDCT images), another for prone position (MR images). Then the corresponding surface meshes were generated using a marching cube algorithm. The next step is to simulate the zero-gravity state of breast from the two models. The bulk modulus of fat and fibroglandular tissue were determined as 3400 Pa and 50000 Pa, respectively. An iterative scheme was applied3. Starting with the gravity loaded models, the same gravity with inverted direction was applied to two meshes. The vertices in the posterior border of the model were fixed, which was configured as the boundary conditions. All simulations were performed with the open source package niftysim4, a Total Lagrangian Explicit Dynamic Solver (TLED). After getting the zero-gravity model n(0), we applied the gravity in the original direction. The difference el was calculated by subtracting the original nodal position and the simulated nodal position. Then el was transformed to unloaded configuration er=F-1el , where F is the deformation gradient. Then zero-gravity model was updated as n(i+1)=n(i)+αer , where α is a scaling factor. Here we used α = 0.65. Once er is smaller than a threshold, the iteration stopped and n(i) was considered as the final unloading gravity state. Next, using the gravity-unloaded supine and prone models, the rigid alignment was applied. Then the Demon’s algorithm was applied to do the final non-rigid registration.Result
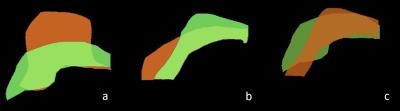
The method was applied to register the MRI and CT of 14 normal volunteers without any symptoms. In Fig. 1, on the original non-fat saturated T2-weighted images, fibroglandular tissue is shown as brighter part and fatty tissue is shown as darker part. After normalization, the intensities of MR images are inversed and adjusted into the same scale as the LDCT images. By this way, the two unloaded models can be used for the intensity-based registration. In Fig. 3, by comparing the original and simulated images, the deformations of both models can be visualized. One example of co-registration using clinical MRI and LDCT images is shown in Fig. 4. The size of the breast on MRI and CT becomes similar after unloading the gravity from supine to prone position, and vice versa.Discussion
We present a method to co-register the breasts in the prone position and the supine position. An iterative algorithm was utilized to unload the gravity, then the unloaded supine and prone images were co-registered using the Demon’s algorithm. The material properties used in the simulation in this work were estimated, which cannot be accurately measured. Therefore, this may be a limitation for the biomechanical simulation method. As breast tissue is deformed, the material properties may change, which cannot be accurately estimated during the simulation process. Nonetheless, we have demonstrated the potential of the proposed method for co-registration. Images acquired using different breast imaging modalities (mammography, ultrasound, CT, MRI, PET, etc.) may be transformed and co-registered, which may provide a tool for future clinical application in diagnosis or treatment planning.Acknowledgements
This work was supported in part by NIH/NCI grants R01 CA127927, R21 CA170955 and R03 CA136071.References
1. Jeon-Hor Chen, Siwa Chan, Nan-Han Lu, Yifan Li, Yu Chieh Tsai, Po Yun Huang, Chia-Ju Chang, Min-Ying Su, Opportunistic Breast Density Assessment in Women Receiving Low-dose Chest Computed Tomography Screening, Academic Radiology, Volume 23, Issue 9, September 2016, Pages 1154-1161, ISSN 1076-6332
2. Lin M, Chen J-H, Wang X, Chan S, Chen S, Su M-Y. Template-based automatic breast segmentation on MRI by excluding the chest region. Medical Physics. 2013;40(12):122301. doi:10.1118/1.4828837.
3. Eiben B, Vavourakis V, Hipwell JH, et al. Symmetric Biomechanically Guided Prone-to-Supine Breast Image Registration. Annals of Biomedical Engineering. 2016;44:154-173. doi:10.1007/s10439-015-1496-z.
4. Johnsen SF, Taylor ZA, Clarkson MJ, et al. NiftySim: A GPU-based nonlinear finite element package for simulation of soft tissue biomechanics. International Journal of Computer Assisted Radiology and Surgery. 2015;10(7):1077-1095. doi:10.1007/s11548-014-1118-5.
Figures

Figure 1.The segmented right breast in a 71-year-old woman. (a)(c): original shape of breast. (b)(d): deformed shape of breast. (a)(b): the original intensity level. (c)(d): the normalized intensity level.