4907
Free-breathing multi-slice ultra-fast SSFP acquisitions for multi-volumetric morphological and functional lung imaging1Department of Radiology, Division of Radiological Physics, University of Basel Hospital, Basel, Switzerland, 2Department of Biomedical Engineering, University of Basel, Basel, Switzerland
Synopsis
Multi-volumetric breath-hold imaging was recently proposed for respiratory $$$\alpha$$$-mapping, a novel quantitative measure of pulmonary ventilation. In this work, we evaluate the feasibility to derive 3D pulmonary functional maps from morphological lung data which are reconstructed from a time-series of multi-slice 2D ultra-fast SSFP scans acquired in free-breathing (4D-MRI).
Introduction
Recently $$$\alpha$$$-mapping was introduced as a novel proton-based ventilation-weighted quantitative measure1, derived from the naturally occurring signal intensity (SI) modulations of the lung at diverse inspiratory phases. In contrast to e.g. Fourier decomposition2, respiratory α-mapping takes into account not only breathing-related SI modulations, but also associated lung volume changes. From a physical lung model relating the observed SI changes to the lung volume1, the derived α-index becomes virtually independent on the inspiratory phase, breathing amplitude and regional spin-density. For respiratory α-mapping, volumetric ultra-fast steady-state free procession (ufSSFP3) scans are acquired at different breath-hold (BH) positions. This technique has shown promising results for reproducible and sensitive detection of respiratory defects in COPD and cystic fibrosis patients. Heavily respiratory compromised patients, infants or young children, however, might be unable to reliably perform breath-holding maneuvers. Therefore, we propose to derive 3D pulmonary functional maps from multi-volumetric data which are reconstructed from a reordered time-series of multi-slice 2D ufSSFP scans acquired in free-breathing (referred to as 2D-FB).Materials and Methods
Measurement protocol
All measurements were performed on a 1.5T clinical MR-scanner using a
12-channel body array and a 24-channel spine receiver coil in a healthy
volunteer positioned supine.
A contemporary 2D-ufSSFP multi-slice sequence2
was modified for coronal multi-slice imaging using a TE/TR = 0.72/1.50 ms, a flip
angle = 40º (maximal due to SAR constrain), a readout bandwidth of 1953
Hz/pixel, a field-of-view = $$$450\times450\times196$$$ mm3 (in-plane spatial
resolution of $$$3.5\times3.5$$$ mm2, interpolated
to $$$1.75\times1.75$$$, slice-thickness 7 mm), a
parallel imaging factor of 2, yielding 8 images/s (125 ms per slice). In
order to let the magnetization to recover between each slice acquisition, two interleaved
groups of 28 slices each were acquired coronally. The acquisition was repeated 48
times in free breathing, resulting in a nominal scan time of 5 min 30s. Neither
prospective gating nor fixed-slice navigator acquisitions were used.
For comparison, the same 2D multi-slice acquisition was acquired also in breath-hold (2D-BH) with an acquisition time of 7 seconds per chest-image, as well as in 3D in 15 seconds with isotropic resolution of 3.1 mm3 [3D-BH, see Ref. (3) for details]. For respiratory α-mapping, BH scans were performed at 5 different inspiratory levels comprised in the tidal breathing region.
Image post-processing and analysis
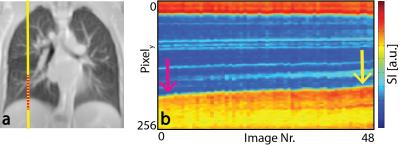
Reconstruction of multi-volumetric data from free-breathing scans was performed with a retrospective respiratory gating. To this end, the diaphragm position was extracted for each slice from a navigator window (cf. Fig. 1). Subsequently, images were binned as a function of the retrospectively derived diaphragm position and recombined to form multi-volumetric datasets at different inspiratory phases (4D-MRI). Finally, respiratory α-maps were derived as previously described1, using $$\alpha\left(\vec{x}\right)=-\frac{\partial\log\left(\textrm{SI}\left(V,\vec{x}\right)\right)}{\partial\log\left(V\right)}\quad[1]$$
after lung volume segmentation, 3D-deformable image-registration (Elastix4), and median filtering5. Generally, $$$\alpha\left(\vec{x}\right)=1$$$ indicates that a region-of-interest (ROI, at position $$$\vec{x}$$$) expands equally to the whole lung mean expansion, whereas $$$\alpha\left(\vec{x}\right)<1$$$ indicates that the ROI expands less than the whole lung mean expansion [for $$$\alpha\left(\vec{x}\right)>1$$$, more].
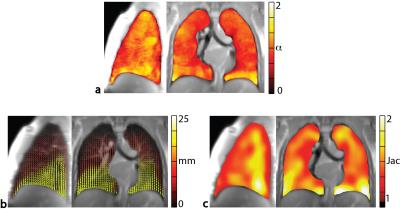
In addition, from the vector-transformations resulting from image co-registration, lung deformation fields and Jacobian-based estimates of lung regional ventilation were calculated.
Result
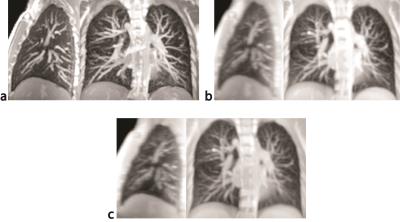
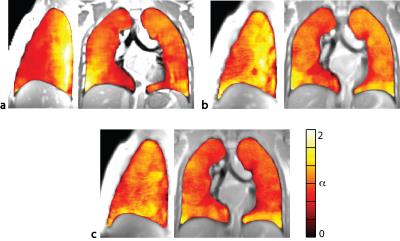
A comparison between 3D-BH, multi-slice 2D-BH and multi-volumetric data reconstructed from multi-slice 2D-FB scans is presented in Fig. 2 in sagittal and coronal views and for a respiratory volume in the middle of the tidal respiratory phase. Overall, despite of the simplistic reordering scheme, only residual misalignments of vessels and diaphragm are recognizable, particularly in the dorsal part of the chest.A side-by-side comparison of respiratory α-mapping resulting from the three acquisition-modalities is presented in Figure 3. Respiratory α-maps are characterized by iso-gravitational homogeneity and a physiological ventral-to-dorsal respiratory-gradient, which is less pronounced for 2D acquisition, especially in FB; we theorize that this effect is caused primarily by non-perfect image reconstruction (reordering) and in second place by physiological effects.
Figure 4 shows respiratory α-maps, deformation field and Jacobian determinant-maps resulting from 2D-FB acquisitions. A moderate visual similarity is seen for the three functional imaging measures, i.e. grater expansion in the inferior lobes.
Discussion and Conclusions
In this work, we have shown the feasibility to obtain 3D functional pulmonary mapping ($$$\alpha$$$-mapping, deformation fields / motion-dynamics and Jacobian-expansion) from multi-volumetric morphological images of the lung which are reconstructed from retrospectively binned multi-slice 2D-FB scans. The reconstructed lung volumes still suffer from small residual misalignments, to be addressed in future investigation, e.g. according to (6).
In conclusion, with an overall acquisition time of approximately 5 minutes, the proposed scheme seems promising for multi-volumetric morphological imaging and for functional respiratory mapping in infants or pediatric patients, as well as in subjects unable to perform breath-holding maneuvers.
Acknowledgements
This work was supported by a grant from the Swiss National Science Foundation (Grant No. 320030_149576).References
[1] Pusterla O, Bauman G, Wielputz MO, Nyilas S, Latzin P, Heussel CP, Bieri O. Rapid 3D in vivo 1H human lung respiratory imaging at 1.5 T using ultra-fast balanced steady-state free precession. Magn. Reson. Med. 2016.
[2] Bauman G, Bieri O. Matrix pencil decomposition of time-resolved proton MRI for robust and improved assessment of pulmonary ventilation and perfusion. Magn Reson Med. 2016 Jan 12.
[3] Bieri O. Ultra-fast steady state free precession and its application to in vivo 1H morphological and functional lung imaging at 1.5 tesla. Magn. Reson. Med. 2013;70:657–663.
[4] Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. Elastix: A toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 2010;29:196–205.
[5] Bieri O. A Method for Visualization of Parenchyma and Airspaces from 3D Ultra-Fast Balanced SSFP Imaging of the Lung at 1.5T. In: Proc. Intl. Soc. Mag. Reson. Med. 22; 2014. p. 2300.
[6] Tong Y, Udupa JK, Ciesielski KC et al. Retrospective 4D MR image construction from free-breathing slice Acquisitions: A novel graph-based approach. Med. Image Anal. 2017;35:345–359.
Figures