4826
A Novel Retrospective Sorting Strategy Utilizing both Respiratory Profile and Internal Surrogate Position for Generating Free-Breathing Abdominal 4D-MRI1Medical Physics and Research Department, Hong Kong Sanatorium & Hospital, Happy Valley, Hong Kong, 2Department of Radiotherapy, Hong Kong Sanatorium & Hospital, Happy Valley, Hong Kong
Synopsis
Four-dimensional (4D) image are widely used to capture the respiration-induced motion of the abdominal organs in free-breathing radiotherapy. However, due to the breathing variability, appropriate assignment of images to each phase is critical in revealing the motion of anatomy structures. In this study, we proposed a novel approach to develop high-quality retrospective 4D-MRI. Instead of sorting images only based on their respiratory phases, the proposed strategy sorted the images using both the respiratory phases and the internal surrogate positions. The proposed strategy was tested using in-vivo human abdominal images.
Purpose
Four-dimensional (4D) image are widely used to capture the respiration-induced motion of the abdominal organs in free-breathing radiotherapy. As an alternative to 4D-CT (1), 4D-MRI technique has demonstrated great potential owning to its high soft-tissue contrast and non-ionizing radiation. Various 4D-MRI techniques have been proposed (2-7). Among them, retrospective 4D-MRI with sequential acquisition holds promise to capture the abdominal respiratory motion at relatively high spatial/temporal resolution (6). In the sequential 4D-MRI, multi-slice MR images are acquired to cover the target volume and the acquisition process is repeated to ensure adequate sampling of all respiratory phases at each slice position. However, due to the breathing variability and restriction of image temporal/spatial resolution, appropriate assignment of images to each respiratory phase is critically important but challenging to faithfully reveal the motion of anatomy structures while preserve structural volume consistency. In this study, we proposed a novel approach to resolve these challenges in developing high-quality retrospective 4D-MRI. Instead of sorting images only based on their respiratory phases, the proposed strategy sorted the images using both the respiratory phases and the internal surrogate positions. The proposed strategy was tested using in-vivo human abdominal images.Materials and Methods
Data Acquisition: Ten healthy volunteers were recruited for this study. Following informed consent, images were acquired on a 1.5T MR-sim (Aera, Siemens Healthineers, Erlangen, Germany) using HASTE sequence in the coronal plane using the sequential mode. The image protocols were TR/TE = 500/53ms, voxel size = 2.5 x 2.5 x 4mm3, slices = 50, FOV = 480mm, matrix size = 192 x 192, flip angle = 111º, repetition = 30, temporal resolution ~ 2fps, iPAT (GRAPPA) = 2, partial Fourier factor = 5/8, acquisition time ~ 12:30.
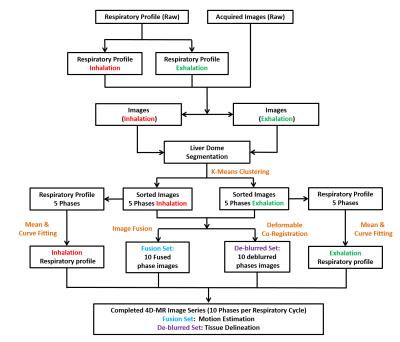
Retrospective Sorting: During the scan, respiratory signal was acquired by using an external surrogate. Each MR image was indexed by its acquisition time and synchronized with the recorded respiratory signal. Then digitized respiratory waveform was categorized into inhalation and exhalation states by calculating the respiratory waveform gradient. Liver segmentation was performed using edge based region growing. To avoid erroneous inclusion of other surrounding tissues, the segmentation was examined and refined manually. Liver dome was considered as internal surrogate in this study and was tracked and classified into 5-phase bins based on its relative position for the inhalation and exhalation states, respectively, using k-means clustering regularized with normalized 2D cross-correlation. At each phase bin, all images were firstly fused on a pixel-by-pixel base to generate a “fusion set” of images to assess the intra-phase anatomic motion. To further minimize the inevitable blurring and generate an artifact-free image set, the “de-blurred set” of images deformedly co-registered all images to form one representative image that is mostly similar to the first-pass image in terms of structure similarity index (SSIM) for each phase bin. The flowchart of proposed retrospective sorting process was shown in Fig.1.
Results and Discussion
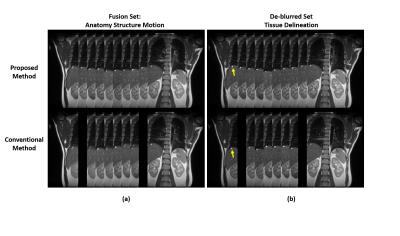
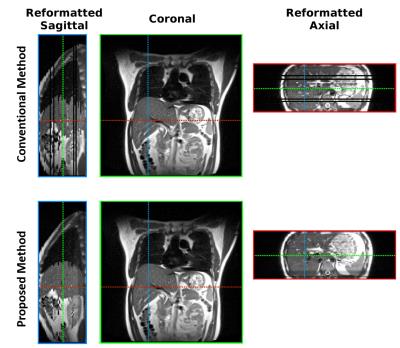
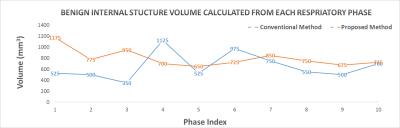
To demonstrate the difference and superiority of the proposed method, we performed a conventional sequential sorting method (6) for comparison. T2-weighted images from 10 respiratory phases of a representative coronal slice are shown in Figure 2. The proposed approach captured the liver dome respiratory variation in a smoother fashion and showed less volume duplicating and volume missing of a benign internal structure (indicated by arrow in Figure 2) compared with the conventional method. The de-blurred image set derived from the proposed method presented sharp and clear tissue structures that would be helpful for accurate tissue delineation. Figure 3 illustrates the reformatted representative sagittal and axial views of the proposed method and conventional method. Better tissue structural continuity and fewer artifacts were achieved by the proposed method, which indicated the more accurate sorting of images to the same respiratory phase. To quantitatively assess the performance of the proposed method, the inter-phase volume change of the internal structure was calculated and plotted in Figure 4. The proposed method considerably reduced the inter-phase volume coefficient of variation (CV) from 35.1% by the conventional method to 18.9%.Conclusion
This work proposed a novel retrospective sorting strategy utilizing both respiratory profile and the segmented liver dome position for generating free-breathing 4D-MRI. Preliminary results showed that the proposed method could achieve accurate respiratory phase sorting and reduce motion-induced tissue volume inconsistency, thus potentially benefiting radiotherapy applications.Acknowledgements
No acknowledgement found.References
1. Ford E. C., Mageras G. S., Yorke E., and Ling C. C., “Respiration-correlated spiral CT: A method of measuring respiratory-induced anatomic motion for radiation treatment planning,” Med. Phys. 2003; 30: 88–97.
2. Tryggestad E, Flammang A, Han-Oh S, Hales R, Herman J, McNutt T, Roland T, Shea SM, Wong J. Respiration-based sorting of dynamic MRI to derive representative 4D-MRI for radiotherapy planning. Med Phys 2013;40(5):051909.
3. Tokuda J, Morikawa S, Haque HA, Tsukamoto T, Matsumiya K, Liao H, Masamune K, Dohi T. Adaptive 4D MR imaging using navigator-based respiratory signal for MRI-guided therapy. Magn Reson Med 2008;59(5):1051-1061.
4. Kuregger S, Nielson T. Efficient repiratory navigator-based 4D MRI. Proc. Intl. Soc. Mag. Reson. Med, 2016; pp 3594.
5. Hu Y, Caruthers SD, Low DA, Parikh PJ, Mutic S. Respiratory amplitude guided 4-dimensional magnetic resonance imaging. Int J Radiat Oncol Biol Phys 2013;86(1):198-204.
6. Liu Y, Yin FF, Rhee D, Cai J. Accuracy of respiratory motion measurement of 4D-MRI: A comparison between cine and sequential acquisition. Med Phys 2016;43(1):179.
7. Stemkens B, Tijssen RH, de Senneville BD, Heerkens HD, van Vulpen M, Lagendijk JJ, van den Berg CA. Optimizing 4-dimensional magnetic resonance imaging data sampling for respiratory motion analysis of pancreatic tumors. Int J Radiat Oncol Biol Phys 2015;91(3):571-578.
Figures