4782
Mixed performance of PIRADS v2: a validation study1Radiology, University of Colorado School of Medicine, Aurora, CO, United States, 2Urology, University of Colorado School of Medicine, Aurora, CO, United States, 3Lake Erie College of Osteopathic Medicine
Synopsis
The second version of the Prostate Imaging Reporting and Data System (PIRADSv2) is the new standard for interpreting prostate MRI. Validation studies have been performed of this scheme, but many have used targeted biopsies as the reference standard. Our validation study is unique in utilizing 3D trans-perineal mapping biopsy (3DTMB) as the reference standard. With a total of 41 MRI lesions, PI-RADSv2 score = 5 lesions had a PPV for cancer of 1.0. However, when all significant lesions (PI-RADSv2 score ≥3) were included, the PPV is 0.29, with equal PPV of PI-RADS 3 and 4 lesions.
Purpose
The second version of the Prostate Imaging Reporting and Data System (PIRADSv2) has become the new standard for interpreting prostate MRI1. Several recent studies have been performed to validate this system2-7. These studies mainly used forms of targeted biopsies without2, 3 and with systematic biopsies4, 6 as the reference standard. Few have used whole mount prostatectomy results as the reference standard5, 7. However, none have used 3D trans-perineal mapping biopsy (3DTMB) as a reference standard. We undertook a study to validate PIRADS v2 by correlating pre-biopsy MRI followed by 3DTMB according to PIRADSv2 criteria for significant lesions (PIRADSv2 category ≥ to 3).
Methods
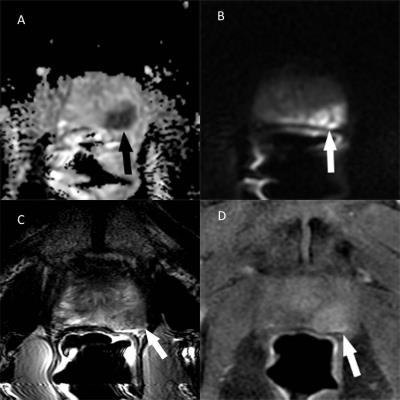
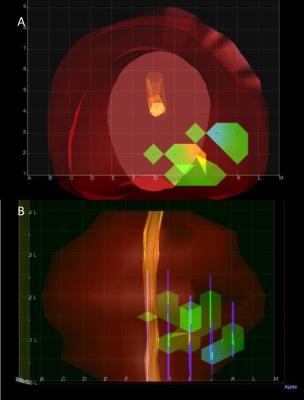
This was an institutional IRB-approved, HIPPA compliant retrospective single institutional cohort study. Patients who were suspected of having prostate cancer and who underwent prostate MRI between September 2010 and June 2013 followed by 3D trans-perineal mapping biopsy were included in the study. MRI was reviewed by two experienced abdominal radiologists independently, with any instances of discordance resolved by a third experienced abdominal radiologist. 3DTMB pathology was independently read by an experienced uropathologist. MRI data were co-registered to the 3DTMB data by dividing prostate gland into 12 segments in both sets of data. We evaluated the association between PIRADSv2 and 3DTMB outcomes using generalized estimating equations regression.
Results
There were a total of 41 MRI lesions called on final consensus in 41 patients. Lesions with PI-RADSv2 score = 5 were significantly associated with cancer and had a positive predictive value (PPV) of 1.0. PIRADSv2 score = 4 or 3 each had a PPV of 0.22, thus scores of 3 compared to 4 were not statistically different. Including all MRI lesions (PIRADSv2 score≥3), the PPV was 0.29.
Discussion
Lesions with a PIRADSv2 score = 5 had a high PPV for prostate cancer on 3DTMP. Interestingly, PIRADSv2 score = 4 lesions have a surprisingly low PPV, which was equal to the PPV for PIRADSv2 score = 3 lesions in our study. The score = 3 and score = 4 data are discrepant from many of the other recent validation studies. In these studies, the reported PPV for PIRADSv2 score ≥3 have been higher, approximately 0.8 4, 6. Other studies did not directly report PPV and used a cutoff of PIRADSv2 score ≥4 but also reported higher accuracy values2, 3, 5.
Conclusion
PIRADSv2 score = 5 lesions have a high positive predictive value for prostate cancer on 3DTMP.
Acknowledgements
No acknowledgement found.References
1. Weinreb, J.C., et al., PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur Urol, 2016. 69(1): p. 16-40.
2. Kasel-Seibert, M., et al., Assessment of PI-RADS v2 for the Detection of Prostate Cancer. Eur J Radiol, 2016. 85(4): p. 726-31.
3. Polanec, S., et al., Head-to-head comparison of PI-RADS v2 and PI-RADS v1. Eur J Radiol, 2016. 85(6): p. 1125-31.
4. Baldisserotto, M., et al., Validation of PI-RADS v.2 for prostate cancer diagnosis with MRI at 3T using an external phased-array coil. J Magn Reson Imaging, 2016. 44(5): p. 1354-1359.
5. Park, S.Y., et al., Prostate Cancer: PI-RADS Version 2 Helps Preoperatively Predict Clinically Significant Cancers. Radiology, 2016. 280(1): p. 108-16.
6. Zhao, C., et al., The efficiency of multiparametric magnetic resonance imaging (mpMRI) using PI-RADS Version 2 in the diagnosis of clinically significant prostate cancer. Clin Imaging, 2016. 40(5): p. 885-8.
7. Greer, M.D., et al., Accuracy and agreement of PIRADSv2 for prostate cancer mpMRI: A multireader study. J Magn Reson Imaging, 2016.
Figures