4779
MR-based machine learning analysis can help to predict pathological outcome of biopsy-proven Gleason score 3+3 prostate cancer1Department of Radiology, the First Affiliated Hospital with Nanjing Medical University, Nanjing,China, People's Republic of China
Synopsis
Machine learning-based analysis of multi-parametric was used to predict pathological outcome of biopsy-proven Gleason score 3+3 prostate cancer and proven to be better compared with single any MR or clinical paremeter in predicting pathological outcome.
Purpose
To investigate whether machine learning-based analysis of multi-parametric (mp) MRI can help to predict pathological outcome of biopsy-proven Gleason score (GS) 3+3 prostate cancer (PCa).Methods and Materials
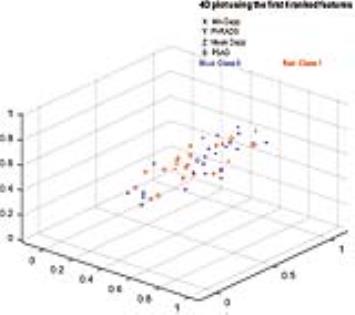
Forty-six patients with biopsy-confirmed GS 3+3 PCa had mp-MRI before prostatectomy. Predicted result was a GS ≥ 3+4 at histopathological findings. Predictive nomogram was established by comprehensive evaluation of clinical and MR features, i.e., age, PSA level, PSA density (PSAD), prostate volume, tumor volume, Dapp and Kapp derived from diffusion kurtosis imaging, PI-RADS v2 score, and MR T-stage, via a novel support vector machine analysis, and tested via area under the receiver operating characteristic curve (Az) analysis.Results
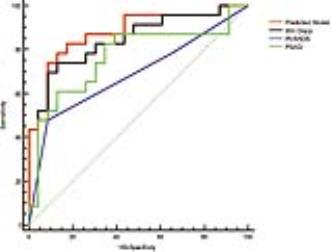
The rate of GS ≥ 3+4 was 50.0% (23/46). Machine learning-based analysis demonstrated that minimum Dapp (weight score: ω = 0.341), PI-RADS score (ω = 0.296), mean Dapp (ω = 0.172) and PASD (ω = 0.107) were the 4 leading ranked features in classifying biopsy outcomes. The developed nomogram had significant higher Az (0.875; 95% confidence intervals [CIs]: 0.744-0.954) than PI-RADS v2 score (Az: 0.687; p = 0.017) and PSAD (Az: 0.764; p = 0.042), while the improvement was insignificant compared to minimum Dapp (Az: 0.832; p = 0.351). Taking same predictive sensitivity, the integral model had significant higher specificity (82.6%) than minimum Dapp (69.6%, p = 0.015), PI-RAD score (34.8%; p < 0.001) and PSAD (65.2%; p = 0.005).
Conclusion
MR-based machine learning analysis can help to predict pathological outcome of biopsy-proven Gleason score 3+3 PCa.Acknowledgements
A part of the data used in this study was previously investigated in our other study with different approach for different purpose.References
No reference found.Figures