4702
Mapping veins on the surface of the human cerebral cortexGünther Grabner1,2, Thomas Haider3, Alexander Rauscher4, Hannes Traxler5, Siegfried Trattnig1, and Simon Daniel Robinson1
1High Field Magnetic Resonance Centre, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 2Department of Radiologic Technology, Carinthia University of Applied Sciences, Klagenfurt, Austria, 3University Clinic for Trauma Surgery, Medical University Vienna, Austria, 4UBC MRI Research Centre, University of British Columbia, Canada, 5Center of Anatomy and Cell Biology, University of Vienna, Austria
Synopsis
Image-guided neurosurgery uses information from a wide spectrum of imaging methods which are registered to the patient's skull so that they correspond to the intraoperative macro- and microscopic view at the start of the operation. During neurosurgical intervention the correspondence between imaging and optical systems breaks because of brain shift down. In this study we demonstrate that Susceptibility-Weighted Imaging and automatic vessel segmentation can be used for visualization and segmentation of superficial cortical veins which can be used as additional reference system during operation.
Purpose:
Image-guided neurosurgery uses information from a wide spectrum of MRI methods to inform the neurosurgeon’s judgement about which tissue to resect and which to spare. For instance, T1-weighted images provide anatomical information and functional MRI activation maps indicate critical and task related populations of neurons. Imaging data from these modalities are registered to the patient's skull so that they correspond to the intraoperative macro- and microscopic view at the start of the operation. During surgery, however, the correspondence between imaging and optical systems breaks down as a result of cerebro-spinal fluid drain, tissue resection, and gravity based brain shift. Currently, the only way to maintain the consistency of imaging and anatomy in terms of brain shift is intraoperative MR imaging which needs a sophisticated setup and is furthermore not widely available. Automatically segmented surface veins could serve as additional reference system with the advantage that they are clearly visible to the surgeon and move and deform with the underlying tissue, which could help to reduce brain shift based location issues [1]. In this study we investigate, using human cadavers, the reliability of visualization and segmentation of superficial cortical veins using Susceptibility-Weighted Images (SWI) [2] and automatic vessel segmentation by reference to high resolution photographs.Materials and Methods:
7 Tesla (Siemens Healthcare, Erlangen, Germany) SWI and T1 weighted imaging was performed on two human cadavers heads using a 32 channel head coil (Nova Medical, Wilmington, USA). SWI was performed using the following parameters: a three-dimensional, fully first-order flow-compensated gradient-echo (SWI) sequence; TE/TR = 10/28 ms; TA=12 min; resolution = 0.3x0.3x1.2mm. Automatic vessel segmentation was performed on the magnitude data using the Frangi vesselness filter [3] which is a multi-scale method that uses the second-order image information, represented by the Hessian matrix to determine the probability that a voxel belongs to a tubular structure (vessel). Within this work the following parameters were used: 0.5 for α and β. C was 1500 and the spatial scale range was between 1.5 and 3. Figure 1 shows the performance of the vesselness filter. For anatomical information, T1-weighted data was automatically segmented using BET (Brain Extraction Tool) [4]. 3D visualization was performed with 3D Slicer and the Medical Imaging Interaction Toolkit (MITK).Results:
Successful vessel segmentation was performed in both cadavers. Figure 1 shows an example of the performance of automatic vessel segmentation on a sagittal brain slice (magnitude data). Surface veins are represented as round, hypointense structures reaching to half of their diameter into the cortex. Segmented vessels are overlaid to the magnitude data in the bottom row of Figure 1. An example of segmented vessels is shown in Figure 2, which represents segmentation results overlaid on anatomical (magnitude) data. As can be seen, veins were successfully segmented from the frontal to the occipital lobe. Figure 2 also gives an impression of the brain shift problem when comparing MRI data to visual appearance on native photographs.Discussion and Conclusion:
Automatically segmented surface veins from SWI correlate with photographs of the surface of the brain, indicating that a mesh of the superficial cortical veins could be used to provide localization information which retains its usefulness during neurosurgical interventions.Acknowledgements
This work was supported by the Österreichische Nationalbank Anniversary Fund project no. 16153 and 16213.References
1) Al-Rekabi et al. Presurgical Visualization of Cerebral Surface Veins with Susceptibility Weighted Imaging, Proc. Intl. Soc. Mag. Reson. Med. 17 (2009; 44) 2) Haacke EM1, Xu Y, Cheng YC, Reichenbach JR.; Susceptibility weighted imaging (SWI).Magn Reson Med. 2004 Sep;52(3):612-8. 3) Frangi, A.F., et al., Multiscale vessel enhancement filtering. Wells WM (ed) III AC F-CaSD, LNCS, 1998: p. 130-137. 4) S.M. Smith. Fast robust automated brain extraction. Human Brain Mapping, 17(3):143-155, November 2002.Figures
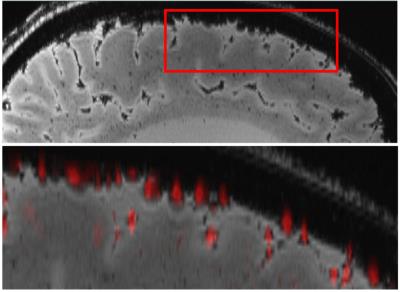
Figure 1: Automatic vessel segmentation: The top row shows a
sagittal brain slice (magnitude), where surface veins can be seen as
hypointense, round structures. The bottom row shows the overlay of the
automatically segmented surface veins (red) with the original magnitude image.
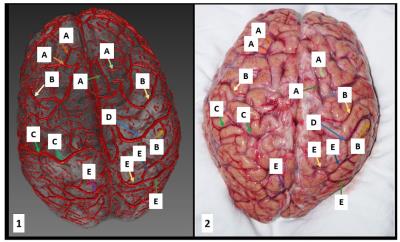
Figure 2: Superficial vein segmentation results overlaid on anatomical
data. Left, the 3D reconstruction and right the corresponding photograph. The labels
represent: A: frontal veins, B: parietal veins, C: the left superior
anastomotic vein (of Trolard), D: the right superior anastomotic vein (of
Trolard), E: occipital veins.