4688
IIT Human Brain Atlas: Enhancement of T1-weighted Template, Tissue Probability Maps and Gray Matter Atlas1Department of Biomedical Engineering, Illinois Institute of Technology, Chicago, IL, United States
Synopsis
The IIT Human Brain Atlas contains anatomical, DTI, HARDI templates, probabilistic gray matter (GM) labels, probabilistic connectivity-based white matter labels, and major
Purpose:
The IIT Human Brain Atlas contains anatomical, DTI1,2 HARDI templates2, probabilistic gray matter (GM) labels3, probabilistic connectivity-based white matter (WM) labels4, and major fiber-bundles of the young adult human brain4. Artifact-free MRI data from 72 human volunteers were used in the development of the atlas5 (diffusion MRI data were collected using Turboprop6). During development of the atlas, spatial normalization was accomplished based exclusively on diffusion data. Consequently, matching of the anatomical information of T1-weighted images was not maximized, blurring the T1-weighted template and tissue probability maps, increasing the noise of the T1-weighted template, and lowering confidence of GM labeling and GM label precision. Therefore, the purpose of this work was to enhance the quality of the T1-weighted template, tissue probability maps, and GM labels of the IIT Human Brain Atlas.Methods:
Data:
The same T1-weighted data from the 72 healthy volunteers included in the previous version of the IIT Human Brain Atlas (v.4.1) were used in the construction of the new resources.
T1-weighted Template:
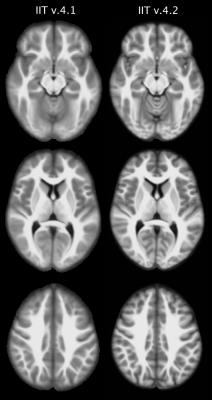
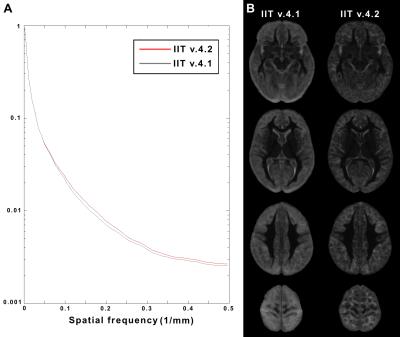
The T1-weighted image-volume of each participant was previously transformed from native space to the space of the existing T1-weighted template2 (v.4.1). These transformed T1-weighted volumes were used in the first step of our approach to construct the new T1-weighted template. More specifically, T1-weighted data from all participants were brought into a common space iteratively, by minimizing both7-9: a) the mean squared intensity difference between each person’s data and a temporary template, and b) the magnitude of all deformations required to map the new temporary template to each participant7-9. In the second step, the spatial transformation generated from the iterative process for each participant was combined with that from native space to the existing T1-weighted template, and the combined transform was applied to the corresponding raw T1-weighted data (single interpolation). In the third step, the T1-weighted data from all participants were averaged to generate the new T1-weighted template (v.4.2). The new and existing templates were compared in terms of sharpness and noise characteristics.
Tissue Probability Maps and Probabilistic Gray Matter Labels:
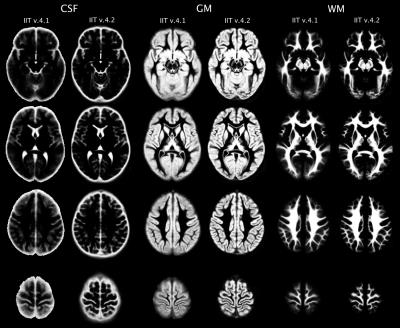
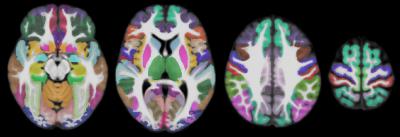
The T1-weighted data of all participants in native space were segmented into GM, WM, and cerebrospinal fluid10 (CSF), and GM was further segmented into 90 cortical and subcortical regions11,12. The tissue masks and GM labels from each participant were then transformed to the space of the new T1-weighted template (v.4.2) using the corresponding transformations generated above. New tissue probability maps were generated and compared to those of the existing atlas (v.4.1). The GM of the new T1-weighted template was divided into 90 cortical and subcortical regions by assigning to each GM voxel a label based on the vote rule. A probability map was generated for each label, separately. A confidence index map was also produced for the GM labeling. The GM labels, probability maps of the labels, and confidence index maps were compared to those of the existing atlas.
Results and Discussion:
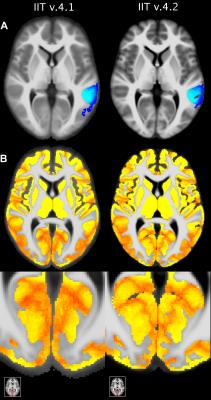
The new T1-weighted template (v.4.2) is of higher sharpness (according to the high spatial frequency content in the corresponding normalized power spectra) (Figs.1,2A), and lower noise than the existing template (Fig.2B). The new tissue probability maps exhibit better definition of GM, WM, and CSF than the existing ones (Fig.3). Figure 4 shows examples of the new GM labels, which generally have probability maps that do not extend far into neighboring labels, and show greater confidence in GM labeling (Fig.5). All improvements presented above are due to the improved spatial matching across participants.Conclusion:
In this work, the T1-weighted template, tissue probability maps and GM labels of the IIT Human Brain Atlas were substantially enhanced. The IIT Human Brain Atlas with its comprehensive set of resources located in the same space, facilitates integration of information from multiple modalities. The improved resources are expected to increase the accuracy of multi-modal studies, as well as conventional investigations on brain macrostructure. The new version of the atlas (v.4.2) is available for download at www.nitrc.org/projects/iit.Acknowledgements
National Institute on Aging R01AG052200References
1. Zhang S, Peng H, Dawe RJ, Arfanakis K. Enhanced ICBM diffusion tensor template of the human brain. Neuroimage 2011;54:974-984.
2. Varentsova A, Zhang S, Arfanakis K. Development of a high angular resolution diffusion imaging human brain template. Neuroimage 2014;91:177-186.
3. Zhang S, Arfanakis K. Development of a Comprehensive Digital Human Brain Atlas. Proc. Int. Soc. for Magn. Reson. in Med. (ISMRM) 2013;p.2129.
4. Varentsova A, Zhang S, Shanina E, Arfanakis K. Connectivity-Based Atlas of Human Brain White Matter in ICBM-152 Space. Proc. Int. Soc. for Magn. Reson. in Med. (ISMRM) 2015.
5. Peng H, Orlichenko A, Dawe RJ, et al. Development of a human brain diffusion tensor template. Neuroimage 2009;46:967-980.
6. Arfanakis K, Gui M, Lazar M. White matter tractography by means of Turboprop diffusion tensor imaging. Ann N Y Acad Sci 2005;1064:78-87.
7. Fonov V, Evans AC, Botteron K, et al. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage 2011;54:313-327.
8. Guimond A, Meunier J, Thirion JP. Automatic computation of average brain models. Medical Image Computing and Computer-Assisted Intervention (MICCAI) 1998;1496:631-640.
9. Guimond A, Roche A, Ayache N, Meunier J. Three-dimensional multimodal brain warping using the demons algorithm and adaptive intensity corrections. IEEE Trans Med Imaging 2001;20:58-69.
10. Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imag 2001;20:45-57.
11. Fischl B. FreeSurfer. Neuroimage 2012;62:774-781.
12. Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006;31:968–980.
Figures