4684
Characterization Of Registration Errors To Screen Aberrant Subject Results Prior To Voxel-Wise Whole Brain Analysis1Gruss Magnetic Resonance Research Center, Albert Einstein College of Medicine, Bronx, NY, United States, 2Radiology, Albert Einstein College of Medicine, Bronx, NY, United States
Synopsis
Voxel-wise analyses of DTI, fMRI or any other MRI-derived metric, be it for comparing groups or one patient to a control group, require quality registration of images to a template. One or several poorly registered images may skew the distributions of the metric in several voxels leading to incorrect inferences. We propose an approach to screen poorly registered images prior to voxel-wise analysis by comparing subject-wise mean displacements of anatomical landmarks between morphed images and the template estimated using FreeSurfer. We apply this proposed algorithm to demonstrate morph accuracy characterization using two age extreme (18 and 86 year old) templates.
Purpose
Voxel-wise analyses of DTI, fMRI or any other MRI-derived metric, comparing groups or one patient to a control group, require quality registration of images to a template. One or several poorly registered images may skew the distributions of the metric in several voxels leading to incorrect inferences. Because quality of registration is sensitive to the similarity of a brain to the template, one method to mitigate this known issue is to use a study-specific "central" template --- a template requiring least average deformation of the brains in the study --- with the idea that smaller deformations are accompanied by smaller registration errors1. Recently, another method to mitigate limitations of registration in patient-versus-group analyses proposed using patient's T1-weighted image as the template because morph errors of the controls average out and are completely eliminated for the patient's data2. However, full error cancellation for controls is not guaranteed, shifting and widening the distributions2.
We propose an approach to screen poorly registered images prior to voxel-wise analysis by comparing subject-wise mean displacements of anatomical landmarks, estimated using FreeSurfer3, between morphed images and the template. We apply this algorithm to demonstrate morph accuracy characterization using two age extreme templates.
Methods
This study was approved by institutional review board of Albert Einstein College of Medicine. The analysis includes T1W images obtained as part of two ongoing studies: Einstein Aging Study (EAS) and Einstein Soccer Study (ESS). Specifically, 108 participants in the EAS cohort are non-demented adults over the age of 70. Exclusion criteria: conditions precluding neuropsychological testing or MRI, active psychiatric symptomatology and non-ambulatory status. Details of the EAS study design are described previously4. The 234 participants of the ESS are adult (age 18-55), active within at least 5 recent years, amateur soccer players, recruited through soccer leagues, clubs and colleges. Exclusion criteria: psychiatric or neurologic disorders. All images were reviewed by an experienced neuroradiologist and determined to be grossly normal brains without visible structural abnormalities.
The imaging was performed using a 3.0T Philips Achieva TX scanner (Philips Medical Systems, Best, The Netherlands) utilizing its 32-channel head coil and the following protocol: T1-weighted 3D MPRAGE with TR/TE/TI=9.9/4.6/900msec, flip angle 8o, 1mm3 isotropic resolution, 240x188x220 matrix.
The T1W image of the brain of each subject was registered to both young (18 years old) and elderly (86 years old) T1-weighted image template brains (Figure 1) using nonlinear registration module of ART5,6. The young brain represented one of the youngest in the ESS study, the elderly is the EAS "central" template identified from the pool of 17 randomly selected EAS subjects by examining deformations after morphing them to each other similar to Smith et al.1.
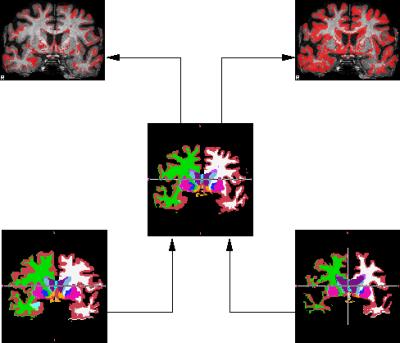
Following registrations, each brain was segmented using the Automatic Subcortical Segmentation of FreeSurfer3. Each atlas was compared to the template's atlas by assigning each voxel a displacement needed to move the voxel to its homologous region, thus producing the map of lower bound of the morph error (Figure 2). In most voxels, the homologous regions overlap and although morph error may be still present it cannot be detected leading to zero lower bound. Each morphed brain was then ascribed a single number: the mean lower bound displacement.
Results
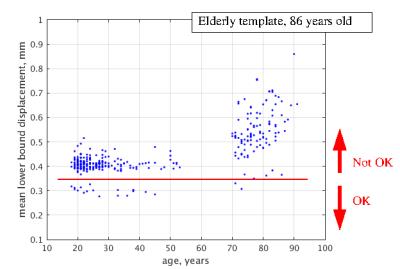
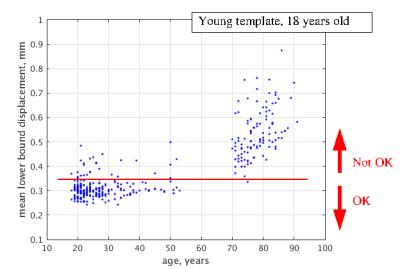
Registration to the EAS’s central template (Figure 3) exhibits large displacements and scatter of the elderly brains despite the template is central and rather compact displacements of the young. This suggests that morph algorithm is not able to deal with variation due to brain atrophy as efficiently as with inherent morphological variation. Only 20 of the brains (3 elderly) show mean lower bound of less than 0.35 mm and represent a well morphed cohort. On contrary, 204 of the brains (1 elderly) register well to the young template (Figure 4) with brains of the elderly demonstrating larger displacements and dispersion.Discussion
Voxel-wise analyses rely on registration to ensure that homologous brain regions are being compared. However, registration processes are limited in the degree to which the anatomy of one brain can be mapped on to another 5-7. Previously, errors in landmark alignment were used to select an overall superior morphing algorithm5-7. We propose error as a metric to assess, screen and filter poorly registered images in single subject or group analyses. Because brain atrophy due to normal aging is known to cause difficulty in registration, we tested this approach by morphing 342 brains across a wide age-span (18-91years) to both young and elderly healthy brain templates and demonstrate a quantifiable metric of morph error associated with age-related atrophy.Conclusion
Our approach quantifies subject-wise registration errors to different templates.Acknowledgements
This work is supported in part by NIH grants 2P01AG003949-32 and 5R01NS082432-05.References
1. Smith S. M., Jenkinson M., Johansen-Berg H., et al. Tract-Based Spatial Statistics: Voxelwise Analysis of Multi-Subject Diffusion Data. NeuroImage 2006; 31: 1487
2. Suri A. K., Fleysher R, Lipton M. L. Subject Based Registration for Individualized Analysis of Diffusion Tensor MRI. PLoS ONE 10(11): e0142288. doi:10.1371/journal.pone.0142288
3. Fischl B., Salat D. H., Busa E., et al. Whole Brain Segmentation: Automated Labeling of Neuroanatomical Structures in the Human Brain. Neuron 2002; 33: 341
4. Katz M. J., Lipton R. B., Hall C. B., et al. Age-Specific and Sex-Specific Prevalence and Incidence of Mild Cognitive Impairment, Dementia, and Alzheimer Dementia in Blacks and Whites: A Report from the Einstein Aging Study. Alzheimer. Dis. Assoc. Disord. 2012; 26: 335
5. Ardekani B. A., Guckemus S., Bachman A., et al. Quantitative Comparison of Algorithms for Inter-Subject Registration of 3D Volumetric Brain MRI Scans. J. Neurosci. Methods 2005; 142: 67
6. Klein A., Andersson J., Ardekani B. A., et al. Evaluation of 14 Nonlinear Deformation Algorithms Applied to Human Brain MRI Registration. NeuroImage 2009; 46: 786
7. Grachev I. D., Berdichevsky D., Rauch S. L., et al. A method for assessing the accuracy of intersubject registration of the human brain using anatomic landmarks. Neuroimage 1999; 9: 250.
Figures