4651
Longitudinal Changes of Metabolites Measured by Proton Magnetic Resonance Spectroscopy and Their Correlations with Behavioral Outcomes in a Rat Model of Kainic Acid Induced Spinal Cord Injury1Diagnostic Radiology, University of Louisville School of Medicine, Louisville, KY, United States, 2Neurological Surgery, University of Louisville School of Medicine, Louisville, KY, United States
Synopsis
We measured the profile of major metabolites longitudinally using MRS in spinal cord gray matter of kainic acid (KA) injured rats at both 7 and 14 days post KA administration and correlated their concentrations with functional outcomes assessed by the Basso, Bresnahan, and Beattie (BBB) Open Field Locomotor scores. Our preliminary findings indicate that metabolite concentrations in the lower lumbar spinal cord gray matter, caudal to the injury epicenter, as detected by in vivo 1H-MRS, could be used as injury and recovery biomarkers in SCI animal models.
PURPOSE
Significant neuronal loss, upregulation of gliosis, as well as impaired locomotor activity and sensory function have been found in studies of spinal cord injury using direct injection, in vivo, of kainic acid (KA), an excitotoxic non-NMDA glutamate receptor agonist into the adult rat spinal cord [1,2]. Proton magnetic resonance spectroscopy (1H-MRS) provides the unique capability of detecting metabolite concentrations in vivo. Previously, we had quantitatively evaluated major metabolites as potential MR based biomarkers of neurotoxicity in this model using MRI/MRS 14 days post KA administration [3]. In current study, we measured the profile of major metabolites longitudinally using MRS in spinal cord gray matter of injured rats at both 7 and 14 days post KA administration and correlated their concentrations with functional outcomes assessed by the Basso, Bresnahan, and Beattie (BBB) Open Field Locomotor scores.METHODS
Animal Model: Female Sprague Dawley rats (n=10) were separated into two groups (n=4 for sham control and n=6 for 2.0 mM KA administration). All procedures were performed according to the guidelines of the University of Louisville Institutional Animal Care and Use Committee (IACUC). For the KA groups, animals received KA injections bilaterally at lumbar level L2. Four intraspinal injections of 0.5 µl each were administered into the intermediate gray matter separated by 1.2 mm rostrocaudally. MRI and MRS were performed at both 7 and 14 days post-injection. Hindlimb function was assessed pre-injury and weekly post-injury using the BBB Locomotor Scale.
MRI/S: All data were acquired using Agilent 9.4 T horizontal bore MRI system equipped with Agilent 205/120 HD gradient coil. A surface coil was carefully positioned on the back of each animal to cover the T10 – L4 spinal cord levels, which included the KA injury site and the more caudal lumbar levels (L3-L4). T2-weighted images were obtained using a standard spin echo multi-slice (SEMS) imaging sequence and then followed by a localized MRS data acquisition. A voxel with a dimension of 1.7 x 2.2 x 6.0 mm was positioned caudal to the KA injury epicenter, aligned with the long axis of the spinal cord. Localization by Adiabatic SElective Refocusing (LASER) sequence was used with the following parameters: TR/TE = 1500/37 msec; spectrum width = 4006 Hz; complex points = 2048; and number of averages = 512. Variable power and optimized relaxation delays (VAPOR) was applied as water suppression scheme. The average total scan time for a single voxel MRS scan was 13 min. MRS data were loaded into jMRUI (version 5.0, UCBL, France) for spectroscopy quantification. Full FID was modeled using AMARES plugin, and major metabolite peak areas were calculated for N-acetyl-aspartate (NAA); total creatine (Cr); total choline (Cho); myo-inositol (mIn), and Glx (glutamine + glutamate at 3.75 ppm).
RESULTS
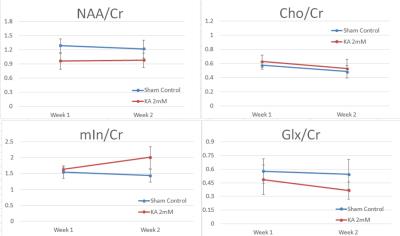
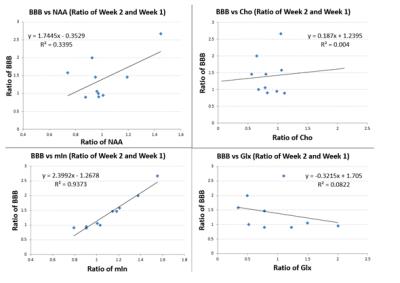
Significantly elevated mIn was observed at two weeks but not at one week post injury comparing to sham control (Fig. 1, p = 0.05). A down-regulation of NAA, a biomarker for neuronal loss, was observed in the KA injury group at both time points. No statistical differences were found in total choline (Cho) at either time point. Glx was lower in the KA injury group, especially at two weeks post injury (Fig. 1). However, when data were assessed as a ratio (week 2/week 1), the changes in NAA and mln were found to be correlated with changes in BBB scores (R= 0.58 and 0.97 respectively, Fig. 2). No correlations were observed between changes in BBB scores and changes of Cho or Glx (Fig. 2).CONCLUSION
Longitudinal evaluation of metabolites in lumbar spinal cord using 1H-MRS in a KA injured rat model was successfully performed at week 1 and week 2 post injury. Our preliminary findings indicate that metabolite concentrations in the lower lumbar spinal cord gray matter, caudal to the injury epicenter, as detected by in vivo 1H-MRS, could be used as injury and recovery biomarkers in SCI animal models.Acknowledgements
No acknowledgement found.References
[1] Magnuson DS, et al., Exp Neurol. 1999 Mar;156(1):191-204. [2] Magnuson DS, et al., Neuroreport. 2001 Apr 17;12(5):1015-9. [3] Zhu M, et al., ISMRM 2016 Proceedings, Abstract 4139