4556
Investigation of machine learning techniques in preoperative glioma grading based on multi-parametric MRI data1Department of Radiology, Tangdu Hospital, Fourth Military Medical University, Xi'an, People's Republic of China
Synopsis
This study demonstrates the significance of integrating multi-parametric MRI attributes and effective machine learning techniques in preoperative glioma grading. A comprehensive scheme combining tumor attribute extraction, attribute selection and classification model was proposed and tested. The tumor attributes were collected from histogram and texture analysis of multi-parameter MRI maps within the whole tumor. The classification performances of 25 commonly used classifiers combined with 8 kinds of attribute selection strategies in differentiating low grade gliomas from high grade gliomas were investigated. Support vector machine (SVM) combined with SVM-RFE attribute selection method were found to exhibit superior performance to others.
Purpose
Preoperative glioma grading is crucial for making treatment plans and improving the prognosis. Focusing on the intrinsic demerits of invasive pathological examination, clinical researchers devoted to investigating a non-invasive glioma grading tool using various magnetic resonance imaging (MRI) techniques1-3. Kinds of quantitative parameters can be derived from multi-modal MRI data to reflect diverse functional features of the brain, which affords the opportunity to explore an automated glioma grade prediction tool utilizing current machine learning techniques4, 5. However, none of previous studies inspected the performance of different types of machine learning models in glioma grading. Thus, a comprehensive glioma grading scheme integrating multi-parametric features with varied machine learning methods was proposed and tested in this study, aiming to establish an effective computer-aided glioma grading model for clinical diagnosis.Methods
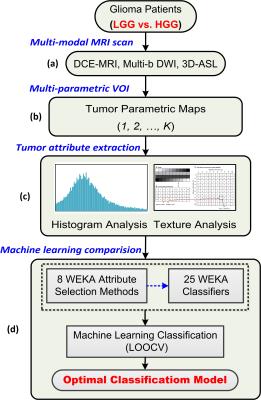
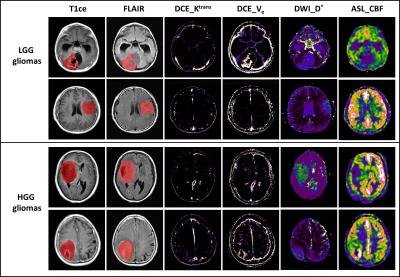
120 histologically confirmed glioma patients were enrolled, involving 28 low grade gliomas (LGGs) and 92 high grade gliomas (HGGs). All of them underwent preoperative multi-modal MRI scans on a 3.0T MRI scanner (MR750, GE Healthcare), approved by the ethical committees of Tangdu Hospital, Fourth Military Medical University. Conventional MRI sequences included axial T1-weighted spin-echo images (T1WI) and contrast enhanced T1WI (T1ce), T2-weighted fast spin-echo images and fluid attenuated inversion recovery (FLAIR). Advanced MRI scans included dynamic contrast enhanced MRI (DCE-MRI), 3-D arterial spin labeling (3D-ASL) and multi-b values diffusion weighted imaging MRI (DWI-MRI) in transverse places. The overall analysis scheme was depicted in Fig. 1. Using the NordicICE software (Version 4.0) and GE post-processing platform, a set of permeability, diffusion and perfusion related parameter maps were generated from DCE-MRI (e.g. Ktrans, Kep, Ve, Vp and etc., 24 parameters), 3D-ASL (e.g. CBF, 1 parameter) and multi-b values DWI-MRI (e.g. D, D* and etc., 5 parameters), respectively (Fig. 2). All of the MRI images were co-registrated into DCE space to retain as much original parameter values as possible. The volume of interest (VOI) covering the whole tumor while excluding the obvious necrosis and edema, was manually drawn on T1ce or FLAIR images, and subsequently overlapped on each parameter image. Then, pixel-by-pixel histogram analysis and first-/second-order texture analysis was performed on each parametric VOI so as to form the big tumor attribute combination (more than 1000 attributes). They were normalized among individuals. An oversampling technique called synthetic minority over-sampling technique (SMOTE) was further applied to reduce the influence of the class imbalance. After that, the classification performances of 25 commonly used classifiers combined with 8 kinds of attribute selection strategies to differentiate LGGs from HGGs were investigated using WEKA (version 3.8.0) software6. The detailed name of each WEKA classifier was summarized in Table 1. Seven attribute ranking strategies were selected, i.e. ‘CorrelationAttributeEval’, ‘GainRatioAttributeEval’, ‘InfoGainAttributeEval’, ‘OneRAttributeEval’, ‘ReliefFAttributeEval’, ‘SymmetricalUncertAttributeEval’, and ‘SVMAttributeEval’ in WEKA, combined with ‘Ranker’ search method. With a stepwise of 50 attributes in each ranking sequence, the optimal attribute subset was determined when the highest classification accuracy was achieved. The last method is named ‘CfsSubsetEval’, which will automatically select the optimal attributes and runs with ‘BestFirst’ method. A leave-one-out cross validation (LOOCV) strategy was applied to assess the performance of each classifier, which is widely used in machine learning studies.Results
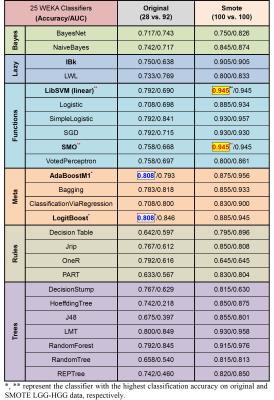
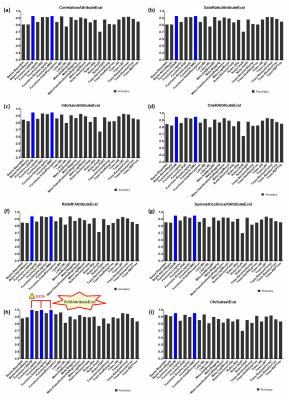
SMOTE technique significantly increased the classification accuracy and AUC compared to using original samples (Table 1). LibSVM, SMO, IBk, SGD, simpleLogistic, LMT and RandomForest classifiers showed superior classification performance to other classifiers when using the whole tumor attribute combination. The best result was acquired using LibSVM and SMO, both of which are support vector machine (SVM) classifiers. When combined with varied attribute selection strategies, SVM classifiers keeps standing the first on the list. Furthermore, the most excellent performance emerged when using ‘SVMAttributeEval’, i.e. SVM Recursive Feature Elimination (SVM-RFE) attribute selection method reported in literature (Fig. 3).Discussion
Multi-parametric MRI maps as well as various histogram and texture statistical indictors were adopted in this study, thus resulting in a big collection of tumor attributes. They may contain much redundant information for classification, which strengthens the importance of attribute selection. Different attribute subsets were picked while using different evaluation strategies. Thus, it is difficult to determine the most critical or relevant attributes for glioma grading. It should be jointly investigated with the classifiers. Besides, a LOOCV strategy was utilized here which took use of the whole data in training and may expand the model’s prediction ability to a certain extent. Therefore, more datasets should be introduced to validate the performance of our classification models.Conclusion
It is concluded that SVM is a promising tool in developing automated preoperative glioma grading system , especially when combined with SVM-RFE attribute selection strategy.Acknowledgements
No acknowledgement found.References
1. Law M, Yang S, Wang H, et al. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am J Neuroradiol, 2003; 24(10):1989-1998.
2. Li X, Zhu Y, Kang H, et al. Glioma grading by microvascular permeability parameters derived from dynamic contrast-enhanced MRI and intratumoral susceptibility signal on susceptibility weighted imaging. Cancer Imaging, 2015; 15(1):1-9.
3. Hu Y C, Yan L F, Wu L, et al. Intravoxel incoherent motion diffusion-weighted MR imaging of gliomas: efficacy in preoperative grading. Scientific Reports, 2013; 4:7208-7208.
4. Svolos P, Tsolaki E, Kapsalaki E, et al. Investigating brain tumor differentiation with diffusion and perfusion metrics at 3T MRI using pattern recognition techniques. Magn Reson Imaging, 2013; 31(9):1567-1577.
5. Fehr D, Veeraraghavan H, Wibmer A, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A, 2015; 112(46): 6265-6273.
6. Zacharaki E I, Kanas V G, Davatzikos C. Investigating machine learning techniques for MRI-based classification of brain neoplasms. International Journal of Computer Assisted Radiology and Surgery, 2011; 6(6): 821-828.
Figures