4510
Localization of Subcortical Structures with the Presence of Lesions in Clinical Brain MRI1Penn Image Computing and Science Laboratory (PICSL), Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States, 2Bioengineering, University of Pennsylvania, Philadelphia, PA, United States, 3School of Information Science and Engineering, Shandong Normal University, Jinan, People's Republic of China, 4Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States, 5School of Computer Science & Engineering, University of Electronic Science and Technology of China, Chengdu, People's Republic of China
Synopsis
In this study, we proposed a pipeline to locate subcortical structures in patients with deep gray matter lesions using clinical brain MRI images. Due to altered signal intensity profile caused by lesions and high slice thickness (~5mm), segmentation of clinical MRI images provide challenges for state-of-the-art algorithms. Our proposed pipeline generates better subcortical structure segmentations, including better lesion coverage and more reliable segmentations than other widely used algorithms. The proposed pipeline may have help in automating the diagnosis of subcortical lesions, potentially improving current clinical practice.
Purpose
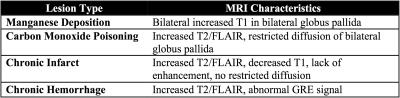
Differential diagnosis of subcortical abnormalities using clinical MRI requires identification of signal changes in various MRI modalities within particular subcortical structures, which include caudate, putamen, pallidum and thalamus. Table 1 lists examples of clinical entities affecting subcortical structures, and their main MRI characteristics. Currently, this task is performed manually by radiologists, which is labor-intensive, time-consuming, and highly subjective. Thus, automatic quantification of MRI signal changes in subcortical structures has the potential to improve clinical practice. To achieve this goal, the first step is to automatically segment subcortical structures from the clinical MRIs, which has distinctive challenges: (1) existing automated algorithms are optimized for research-purpose MRIs (~1mm slice thickness) and often fail due to the high slice-thickness of clinical MRIs (~5mm); (2) state-of-the-art algorithms commonly make assumptions about the signal and location of subcortical structures, which may be violated in the presence of lesions. In this study, we created a pipeline for localizing subcortical structures optimized for clinical MRI accounting for the presence of lesions. The segmentation quality of the proposed method was compared to state-of-the-art algorithms.Materials and Methods
Participants and MRI Protocol: 73 patients who were diagnosed at the Hospital of the University of Pennsylvania with clinical entities primarily affecting subcortical gray matter (GM) structures were included in this study. Patients with significant lesions outside the deep gray nuclei were excluded. Each imaging data set consisted of up to seven different MR sequences, which include axial/sagittal T1-weighted (T1W), T2-weighted, FLAIR, T1W post-contrast, susceptibility-weighted gradient echo (GRE), high b-value diffusion-weighted images (DWI) and apparent diffusion coefficient (ADC) maps. The MR images are clinical grade with ~5mm slice thickness and ~0.5mmx0.5mm in-plane resolution.
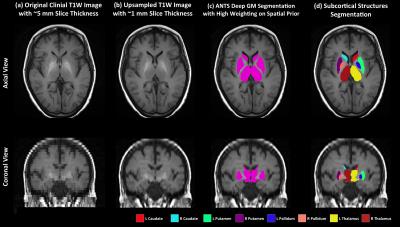
T1W Image Processing: Subcortical structures, including bilateral caudate, putamen, pallidum and thalamus were segmented using the axial T1W image. In cases where axial T1W images were not available (11 cases), the sagittal T1W images were used instead. The T1W image (Figure 1a) is first upsampled to reduce the slice thickness to ~1mm (Figure 1b) using a super-resolution technique1, which has been shown to outperform traditional interpolation approaches. The ANTS cortical thickness pipeline2 was then applied to perform tissue segmentation (including cortical GM, white matter, cerebrospinal fluid, deep GM, brain stem and cerebellum) with high weighting of the spatial prior (the weight is 0.75, 0.25 is the default value), and registered to the OASIS template3. The rationale of using a high spatial prior weight is (1) to give more emphasis on the expected location of the subcortical structures and (2) to allow a larger range of intensity fluctuation of those structures. In order to separate the subcortical structures from one compound label of deep GM generated from ANTS (Figure 1c), the AAL brain parcellation4 in the template space is first warped to the subject T1W space and each deep GM voxel is assigned to the closest subcortical label in the warped AAL parcellation. In the end, morphometry correction is applied to generate the final segmentation (Figure 1d). For comparison, FreeSurfer5 and FIRST6 were applied to the upsampled T1W image to generate subcortical structure segmentations.
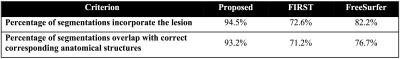
Evaluation: Since our goal was detecting the presence of lesions in addition to generating accurate segmentations, segmentation success was defined according to the following two criteria: (1) the segmentation incorporated the lesion; (2) segmentation labels qualitatively overlapped with the correct corresponding anatomical structures. The evaluation was performed visually by radiologists.
Results and Discussion
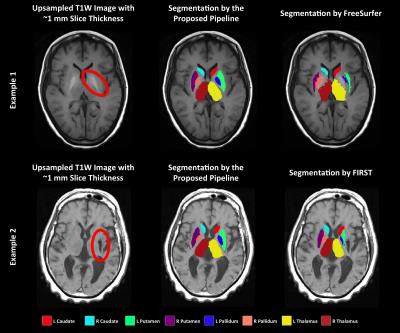
Table 2 shows the result of the comparisons between the proposed pipeline, FreeSurfer5 and FIRST6. Notably, only 72.6% of FIRST segmentations incorporate the lesion and 71.2% of segmentations included labels that overlap well with the corresponding anatomical structure. The FreeSurfer succeeded in a more cases (82.2% and 76.7% correspondingly). The proposed pipeline generated the best results. 94.5% of the segmentations incorporate the lesion and 93.2% of the segmentations are correct. Figure 2 shows two examples where the proposed pipeline generated better segmentations than the other methods. Even though the signal intensity of the structures may deviate significantly from normal GM due to the presence of a lesion, higher weighting of the spatial prior allowed our pipeline to include the lesion in the segmentation in most (>94%) cases.Conclusion
In this study, we proposed a pipeline to accurately locate subcortical structures with the presence of lesions on clinical brain MRIs. Our results indicate that the proposed pipeline generates better subcortical structure segmentations on clinical imaging compared to the state-of-the-art algorithms. As such, the proposed pipeline could have important utility in automatically detecting subcortical abnormalities, a crucial first step in automating the diagnosis of clinical entities involving the basal ganglia.Acknowledgements
This work was supported by National Institute of Health of the United States (R01-EB017255) and National Natural Science Foundation of China (No.61303007).References
1. Manjón J V, Coupé P, Buades A, Fonov V, Collins LD, Robles M. Non-local MRI upsampling. Med Image Anal. 2010;14(6):784-792.
2. Das SR, Avants BB, Grossman M, Gee JC. Registration based cortical thickness measurement. Neuroimage. 2009;45(3):867-879.
3. Marcus DS, Fotenos AF, Csernansky JG, Morris JC, Buckner RL. Open access series of imaging studies: longitudinal MRI data in nondemented and demented older adults. J Cogn Neurosci. 2010;22(12):2677-2684.
4. Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15(1):273-289.
5. Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774-781.
6. Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56(3):907-922.
Figures