4451
Mapping cell shape and cell density by diffusion variance decomposition (DIVIDE): Towards rapid non-invasive diagnostic and prognostic assessment of tumors1Diagnostic Radiology, Lund University, Lund, Sweden, 2Medical Radiation Physics, Lund University, Lund, Sweden, 3CR Development, Lund, Sweden, 4Center for Imaging and function, Skane University hospital, Sweden
Synopsis
In this work we present a novel approach based on tensor-valued diffusion encoding to quantify tumor tissue characteristics such as cell shape and cell density variation — features considered in histopathology. This approach constitutes a promising framework for non-invasive diagnostic and prognostic assessment of tumors that may improve patient care by non-invasively capturing histopathological information on cell shape and cell density heterogenity.
Clinical Question
How can histopathological information on cell shape and cell density heterogeneity in intracranial tumors be obtained preoperatively and non-invasively?Impact
Cancer will be the leading cause of death in the next decade according to WHO. Total cost for cancer care, premature death, and disability is larger than for any other disease.1 In 2012, intracranial brain tumors were the cause of death of 200,000 people worldwide. Additional 250,000 people were diagnosed with intracranial brain tumors on that year.2 High-grade primary brain tumors are amongst the deadliest, with median survival time of 12–14 months and a 10% 5-year survival. Surgery is a cornerstone in the treatment of intracranial tumors. A complete resection and cure are attainable in benign tumors such as meningiomas, and longtime survival is correlated with the extent of resection in high-grade primary brain tumors. MRI is the first step in the routine preoperative workup for intracranial tumors, and is the main tool for visualization of tumor extent and basic characterization of the tumor. However, at present, the level of characterization obtained by MRI is not sufficient by itself for diagnosis and prognosis of tumors. As such, currently, definitive diagnosis is obtained by surgical biopsy. Biopsy is associated with risk for hemorrhage, infection, and seeding. Complications occur in 6–12% of cases.3 Surgical biopsy and resection must be directed towards the most malignant tumor component, warranting comprehensive preoperative characterization of the tumor tissue. It would be extremely beneficial if the characterization could be performed by MRI. This would remove the risk associated with invasive biopsies. Thus it is highly desirable for patient management to extend the current routine MRI with new clinical sequences for the determination of histopathological features.Approach
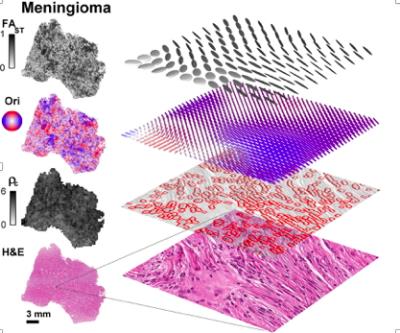
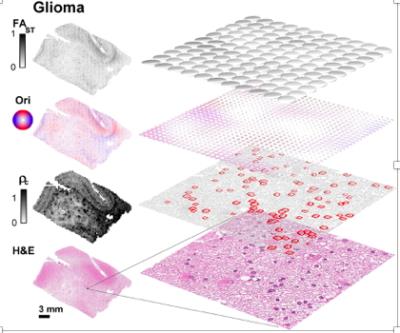
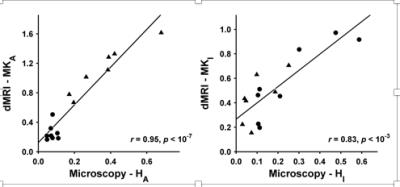
Conventional dMRI is an excellent method for detecting changes of tissue microstructure, but unfortunately it suffers from low specificity, since it cannot distinguish between tumors characterized by different types of cellular microstructural arrangements observed by histopathology. For example, the apparent diffusion coefficient (ADC) obtained with conventional dMRI is an average metric that cannot capture microstructural heterogeneity within individual voxels. Our innovation4 is a multidimensional framework that relies on tensor-valued diffusion encoding to quantify tumor heterogeneity such as cell shape and intra-voxel cell density variation also obtained from histopathology (Fig 1).5 This framework is used for the DIVIDE protocol, which in brain tumors provides parameters that show excellent agreement with corresponding histopathology parameters (Fig 2).5 Cell shape and cell density heterogeneity from histopathology were respectively described by the anisotropic variance (MKA) and the isotropic heterogeneity (MKI). Importantly, and unlike conventional dMRI methods, such as DTI, the MKA parameter from DIVIDE can detect and estimate the diffusion anisotropy even in structures that are disordered, which is frequently the case in tumors.Gains and Losses
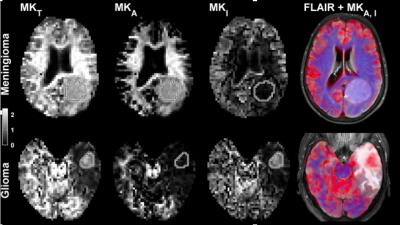
Our results show the abilities of the innovation to provide non-invasive characterization of cell shape and variation in cell density in tumors in good agreement with histopathology.5 In cases where the tumor grade and type are mainly characterized by cell shape and variations in cell density, DIVIDE may improve the diagnostic procedure. For example, meningioma and glioma tumors could be differentiated based on their cell shape (Fig 3).5 We hypothesize that presurgical planning could be improved in meningioma tumors by assessing their toughness from DIVIDE, which cannot be done from conventional dMRI alone.6 Total scan time is less than 8 min, which allows DIVIDE to supplement a standard clinical protocol, to plan and guide the surgical biopsy and/or resection. While DIVIDE provides important characterizations of cell shape and cell density variations, it is limited in its ability to characterize more subtle features such as irregular nuclei shapes and sizes, frequency of mitosis etc. Additional studies are needed to explore the correlation between DIVIDE parameters and different grades and types of intracranial tumors, as well as to validate the sensitivity of DIVIDE for the evaluation of treatment effect, although previous studies suggest that a correlation should exist between tumor grade and diffusional variance7, 8. DIVIDE is comparable to routine dMRI with regards to patient safety.Preliminary data
Our results, published by Szczepankiewicz et al.5, constitute initial evidence of the link between DIVIDE parameters and histopathological measures (Fig 2), which potentially enables non-invasive tumor characterization. The cost for surgical biopsies (a single biopsy costs on average $15000 including hospitalization)2 could be significantly reduced if a fraction of these were replaced with MRI methods such as DIVIDE, which approximately costs $1000-5000. In addition, presurgical planning of the biopsy as well as resection may be improved leading to reduced need of additional biopsies and more efficient tumor resection.Acknowledgements
References
1. John R et al. The Global Economic Cost of Cancer. American Cancer Society and the LIVESTRONG organization (2010).
2. Cancer research UK (http://www.cancerresearchuk.org/home-page).
3. Malone et al. Complications Following Stereotactic Needle Biopsy of Intracranial Tumors. World Neurosurg., (2015).
4. Lasic et al. Microanisotropy imaging: quantification of microscopic diffusion anisotropy and orientational order parameter by diffusion MRI with magic-angle spinning of the q-vector. Front. Physics 2, (2014).
5. Szczepankiewicz et al. The link between diffusion MRI and tumor heterogeneity: Mapping cell eccentricity and density by diffusional variance decomposition (DIVIDE). NeuroImage, (2016).
6. Tropine et al. Differentiation of Fibroblastic Meningiomas From Other Benign Subtypes Using Diffusion Tensor Imaging. J. Magn. Reson. Imaging, (2007)Beckers Hospital Review (http://www.beckershospitalreview.com).
7. Raab et al. Cerebral gliomas: diffusional kurtosis imaging analysis of microstructural differences, Radiology, (2010)
8. Van Cauter et al. Gliomas: diffusion kurtosis MR imaging in grading. Radiology, (2012)
Figures